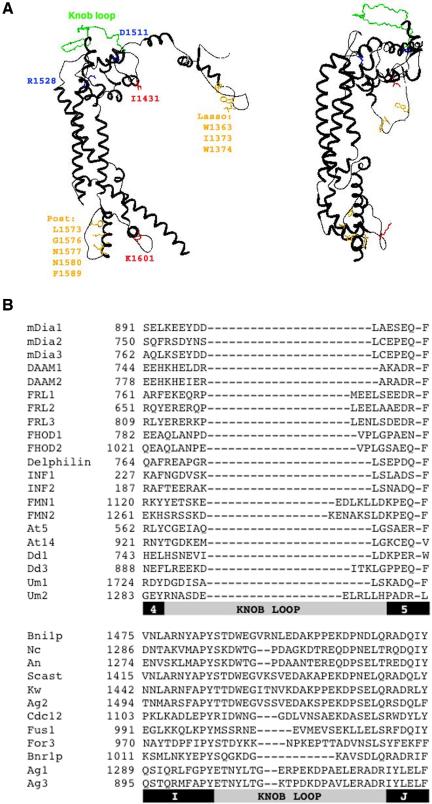
Figure 1.
Summary of FH2 domain alignment results. (A) Models of one subunit from the dimeric Bni1p FH2 domain structure, made using DeepView (www.expasy.org/spdbv/) based on published structure (Xu et al., 2004; PDB file 1UX5). Left, highly conserved residues are shown in orange for dimerization interface (W1363 = 95/101 sequences identical; I1373 = 85/101 for aliphatic and 97/101 for aliphatic or aromatic; W1374 = 97/101 for aromatic; L1573 = 95/101 identity and 99/101 for aliphatic; G1576 = 98/101 identity; N1577 = 100/101 identity; N1580 = 98/101 identity; F1589 = 95/101 as F or Y), red for residues key to effects on actin (I1431 = 93/101 identity and 98/101 as I or V; K1601 = 98/101 as base), and blue for residues important for knob structure (D1511 = 93/101 as acid; R1528 = 100/101 as R or K). The extended knob loop, unique to the Ascomycota fungi, is shown in green. Right, rotated 90° to show the extent to which the Ascomycota knob loop (green) projects over the rest of the knob. (B) Alignment of a segment of the knob region for a selection of FH2 domains. The first set of sequences is from non-Ascomycota, including 15 mouse sequences (to FMN2), one member of each Arabidopsis group, two Dictyostelium sequences, and two Basidomycota sequences. The second set of sequences is from Ascomycota. Refer to Table 1 for sequence abbreviations. Below the alignment, bars depicting helices (black) and the knob loop regions (gray) in the structures of mDia1 and Bni1p, respectively, are illustrated.