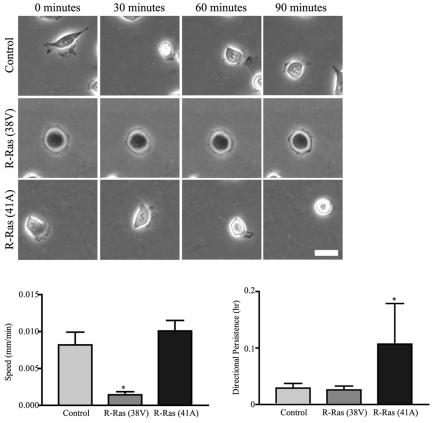
Figure 2.
R-Ras regulates random cell migration across collagen. Control T47D cells, cells stably expressing activated R-Ras (38V), or cells stably expressing dominant negative R-Ras (41A) were plated on collagen-coated Petri plates (3 μg/ml) for 1 h. Time-lapse microscopy sequences were acquired for 90 min, with one image collected per minute (see Supplementary Videos 1, 2, and 3). Shown here is a representative cell at 0, 30, 60, and 90 min during the observation period. Scale bar, 25 μm. Expression of activated R-Ras significantly (*p < 0.05 vs. control) decreases cell speed, whereas expression of dominant negative R-Ras (41A) increases (p = 0.2) speed. Twenty individual cells from three experiments were tracked using a cell tracking program (Nanotrack, Inovision Software) and cell speed (mm/min) was calculated using the random walk equation (Maheshwari and Lauffenburger, 1998). Expression of dominant negative R-Ras (41A) increases directional persistence (*p < 0.05 vs. control).