Abstract
Prohibitins comprise a remarkably conserved protein family in eukaryotic cells with proposed functions in cell cycle progression, senescence, apoptosis, and the regulation of mitochondrial activities. Two prohibitin homologues, Phb1 and Phb2, assemble into a high molecular weight complex of ∼1.2 MDa in the mitochondrial inner membrane, but a nuclear localization of Phb1 and Phb2 also has been reported. Here, we have analyzed the biogenesis and structure of the prohibitin complex in Saccharomyces cerevisiae. Both Phb1 and Phb2 subunits are targeted to mitochondria by unconventional noncleavable targeting sequences at their amino terminal end. Membrane insertion involves binding of newly imported Phb1 to Tim8/13 complexes in the intermembrane space and is mediated by the TIM23-translocase. Assembly occurs via intermediate-sized complexes of ∼120 kDa containing both Phb1 and Phb2. Conserved carboxy-terminal coiled-coil regions in both subunits mediate the formation of large assemblies in the inner membrane. Single particle electron microscopy of purified prohibitin complexes identifies diverse ring-shaped structures with outer dimensions of ∼270 × 200 Å. Implications of these findings for proposed cellular activities of prohibitins are discussed.
INTRODUCTION
In accordance with their strong conservation and widespread distribution, prohibitins have been functionally linked to important cellular processes, such as cellular signaling and transcriptional control (Terashima et al., 1994; Montano et al., 1999; Sun et al., 2004), apoptosis (Vander Heiden et al., 2002; Fusaro et al., 2003), cellular senescence (McClung et al., 1992; Coates et al., 1997; Coates et al., 2001; Piper et al., 2002), early development of Caenorhabditis elegans (Artal-Sanz et al., 2003) or mitochondrial biogenesis (Berger and Yaffe, 1998; Steglich et al., 1999; Nijtmans et al., 2000; Artal-Sanz et al., 2003). However, it remained largely elusive how prohibitins function on the molecular level and whether a similar mode of action underlies these apparently divergent roles of prohibitins.
At least two homologous proteins exist in eukaryotic cells, Phb1 (BAP32, prohibitin) and Phb2 (BAP37, prohibitone, REA), multiple copies of which assemble into a high-molecular-weight complex of ∼1.2 MDa in the inner membrane of mitochondria in yeast, mammals, and C. elegans (Steglich et al., 1999; Nijtmans et al., 2000; Artal-Sanz et al., 2003). In Saccharomyces cerevisiae, both homologues are functionally interdependent: deletion of one subunit results in the destabilization of the other, strongly suggesting that the prohibitin complex represents the functionally active structure (Berger and Yaffe, 1998). Consistently, similar phenotypes are associated with mutations in PHB1 and PHB2. Δphb1 or Δphb2 cells show a shortened replicative life span and an altered mitochondrial morphology in aged cells, pointing to a role during senescence (Coates et al., 1997; Piper et al., 2002). Moreover, deletions of prohibitin genes are lethal in combination with mutations of the inheritance machinery (Berger and Yaffe, 1998) and the phosphatidylethanolamine biosynthetic pathway (Birner et al., 2003) of mitochondria and cause a slow growth phenotype in the absence of the m-AAA protease (Steglich et al., 1999), a key component of the quality control system within the inner membrane of mitochondria. Notably, a supercomplex of prohibitins with the m-AAA protease has been described which link prohibitin function to the turnover of membrane proteins. An accelerated degradation of nonnative polypeptides by the m-AAA protease suggests a regulatory role of prohibitins in this process (Steglich et al., 1999). Moreover, the interaction of nonassembled respiratory chain subunits with the prohibitin complex has lead to the proposal of a chaperone activity of prohibitins during the biogenesis of the respiratory chain (Nijtmans et al., 2000).
Although the function of the prohibitin complex within mitochondria is beginning to emerge, some of the postulated roles of prohibitins in mammalian cells such as transcriptional regulation or cellular signaling are difficult to reconciliate with a mitochondrial location. Even more, Phb1 and Phb2 were found to be exclusively associated with each other in mammalian cells (Coates et al., 2001). On the other hand, mammalian Phb1 and Phb2 have been repeatedly detected also in the cytosol and the nucleus, pointing to potentially different cellular locations under differing conditions (Thompson et al., 2001; Wang et al., 2002; Fusaro et al., 2003; Kurtev et al., 2004; Sun et al., 2004).
In view of the interdependence of Phb1 and Phb2 and the apparently contradicting findings with respect to their cellular location, understanding prohibitin function will require the structural characterization of the prohibitin complex as well as the intracellular targeting of its subunits. We have therefore analyzed the biogenesis of prohibitins in mitochondria of S. cerevisiae, purified the prohibitin complex, and characterized its architecture by electron microscopy.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
Yeast strains used in this study are derivatives of W303-1B. Cells were cultivated at 30°C in YP medium supplemented with 2% glucose or 2% galactose and 0.5% lactate, or in lactate medium. Δphb1 (YGS401) and Δphb2 (YGS501) strains were described previously (Steglich et al., 1999). The Δphb1Δphb2 strain (YGS507) was obtained by deletion of PHB2 in YGS401 by polymerase chain reaction (PCR)-targeted homologous recombination (Wach et al., 1994) by using the heterologous marker cassette KanMX4. Yeast strains expressing C-terminally truncated version of Phb1 or Phb2 were generated by insertion of DNA fragments containing a termination codon and the KanMX4 marker into genomic PHB1 or PHB2 genes by homologous recombination. Termination codons were inserted to replace the base triplets AAC encoding N285 or TTC encoding F181 of PHB1, and the base triplets AGG encoding R308 or TTC encoding F192 of PHB2, generating the strains YTT214 [phb1(Δ284-287)::KAN], YTT216 [phb1(Δ181-287)::KAN], YTT220 [phb2(Δ308-310)::KAN] and YTT222 [phb2(Δ192-310)::KAN], respectively. Integration of the marker cassettes was verified by PCR. The Tim23(fs)strain has been described previously (Sirrenberg et al., 1996). The Tim23(Gal10) strain was kindly provided by D. Mokranjac (University of Munich, Munich, Germany).
Cloning Procedures
For SP6-polymerase-driven expression of PHB1 and PHB2 in vitro, both genes were amplified by PCR and cloned into the BamHI and PstI sites of pGEM4 (Promega, Madison, WI), thereby generating pTT26 and pTT29, respectively. Fragments of PHB1 or PHB2 were cloned into pGEM4 in the same way (pTT27 [Phb1(29-287)], pTT80 [Phb1(1-180)], pTT30 [Phb2(36-310)], pTT31 [Phb2(62-310)], pTT32 [Phb2(1-213)], and pTT33 [Phb2(Δ36-61)]). For the expression of a moiety of Phb1 or Phb2 fused to mouse DHFR, pTT34 [Phb1(1-28)-DHFR], pTT57 [Phb1(1-57)-DHFR], pTT58 [Phb1(1-83)-DHFR], pTT35 [Phb2(1-35)-DHFR], pTT36 [Phb2(1-61)-DHFR], and pTT37 [Phb2(36-61)-DHFR] were generated by cloning of the relevant PCR fragment into a BamHI site in front of the open reading frame encoding mouse DHFR in pGEM4.
For overexpression of Phb1 and Phb2 in yeast, PHB2 and a DNA fragment encoding hexahistidine-tagged Phb1 were amplified by PCR and cloned into the two multiple cloning sites of the pESC-URA vector (Stratagene, La Jolla, CA) allowing the inducible expression from GAL10 and GAL1 promoters, respectively (pTT18). For constitutive expression using the TPI promoter (pTT46(Phb1), pTT47(Phb1His), pTT50[Phb1(1-284)], pTT81[Phb1(1-180)], pTT44(Phb2), pTT54[Phb2(1-303)], pTT55[Phb2(1-216)]), PHB1, and DNA-fragments encoding hexahistidine-tagged Phb1, Phb1(1-284) or Phb1(1-180) were cloned into the NcoI and XhoI-sites of pYX142 (Novagen). PHB2 and variants encoding Phb2(1-303) or Phb2(1-213) were cloned as EcoRI/HindIII-DNA-fragments into pYX132 (Novagen, Madison, WI). PHB2 also was cloned into the EcoRI and HindIII sites of the multicopy vector pYX232 (Novagen), generating pTT45. To obtain strain YTT160 or YTT163, ΔphbΔphb2 cells were transformed with pTT44 and pTT46, or with pTT44 and pTT47, respectively.
Purification of the Prohibitin Complex
Δphb1 (YGS401) cells were transformed with both pTT18 and pTT45. The transformants (YTT51) were grown at 30°C on SC-glucose medium lacking histidine, uracil, and tryptophane. For induction of prohibitin gene expression, logarithmically growing cells were transferred to YP medium supplemented with 2% galactose and 0.5% lactate and further incubated for 6 h at 30°C. Then, cells were isolated, washed with buffer A (20 mM HEPES/KOH, pH 7.4, 20 mM KCl, 2 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], EDTA-free protease inhibitor cocktail [Roche Diagnostics, Indianapolis, IN]) and lysed using two times a Emulsi-flex C5 cell disrupter (Avestin Europe, Mannheim, Germany) at ∼24,000 psi. After two clarifying spins at 1000 × g for 5 min, mitochondrial membranes were isolated by centrifugation at 17,000 × g for 15 min. Mitochondrial membrane proteins were resuspended (4 mg/ml) in buffer B (20 mM HEPES/KOH, pH 7.4, 50 mM K-phosphate buffer, pH 7.4, 150 mM KCl, 5 mM ATP, 5 mM magnesium acetate, 1 mM PMSF, EDTA-free protease inhibitor cocktail [Roche Diagnostics], 10% glycerol, 10 mM imidazole, 1% [wt/vol] n-dodecyl-β-d-maltopyranoside [DDM]) and solubilized by incubation for 20 min at 4°C. Insoluble material was removed by centrifugation at 110,000 × g for 15 min. The supernatant was loaded on Ni2+-conjugated Hi-trap chelating column (1 ml; Amersham Biosciences, Piscataway, NJ). Bound proteins were eluted by a linear gradient of sorbitol in buffer C [20 mM HEPES/KOH, pH 7.4, 50 mM K-phosphate buffer pH 7.4, 150 mM KCl, 1 mM ATP, 1 mM Mg(OAc)2, 1 mM PMSF, 10% glycerol, 0.1% (wt/vol) DDM]. Peak fractions (2 ml) were concentrated twenty times by Centricon YM-100 (Millipore, Bedford, MA), and then subjected to glycerol gradient sedimentation for 3 h at 268,000 × g (10-30% glycerol in buffer C). Fractions were collected from the top and analyzed by SDS-PAGE and silver staining. Fractions containing the prohibitin complex were pooled and examined for the presence of abundant mitochondrial protein complexes (Hsp60, porin, F1FO-ATP synthase, and respiratory chain complexes), mitochondrial translocases (TOM and TIM complexes), i- and m-AAA protease complex, or 26S proteasomes by immunoblotting by using specific antisera against subunits of these complexes.
Single Particle Electron Microscopy of Prohibitin Complexes
Electron Microscopy. Individual glycerol gradient fractions consisting of 10-30% (wt/vol) glycerol in buffer C were applied to freshly glow-discharged 400 mesh carbon-coated copper grids and stained with 1% of phosphotungstate, pH 7.0. Corresponding fractions resulting from purifications by using Δphb1 cells simultaneously overexpressing both Phb1His and Phb2 from one multicopy plasmid as well as Δphb1Δphb2 cells were directly compared with detect any contaminants at background level. Micrographs were recorded in a Philips CM 120 electron microscope operating at 100 kV at a nominal magnification of 45,000× under low electron dose conditions covering a defocus range of 0.8-1.4 μm.
Negatives were digitized on a flat-bed SCAI (Carl Zeiss, Jena, Germany) microdensitometer with 7-μm pixel size and subsequently reduced to a final pixel size of 21 μm corresponding to 4.7 Å on the specimen scale. Particles were manually selected from four micrographs by using WEB software (Frank et al., 1996). After extraction of 303 frames with 128 × 128 pixels, images were corrected for the effects of the contrast transfer function (Radermacher et al., 2001). Translational/rotational alignment was performed using the SPIDER/WEB software (Frank et al., 1996) with extensions. A first reference was created by the reference-free alignment algorithm (Marco et al., 1996) followed by reference-based alignments by using cross-correlation of two-dimensional Radon transforms. After each round of multireference alignment, images were subjected to multivariate statistical analysis (Van Heel and Frank, 1981). Subsequently, class averages were calculated by hierarchical ascendant classification applied to moving cluster centers and then used as references in subsequent rounds of refining particle alignments. At the final stage, 159 node images were selected from neural network analysis implemented in the XMIPP package (Marabini et al., 1996). The resulting data set was split into two subsets after masking and average images were generated from self-organizing maps of each subset.
Miscellaneous
The following procedures were performed essentially as described previously: in vitro import into mitochondria and chemical cross-linking (Arlt et al., 1996) and blue native gel electrophoresis (Schägger, 2001).
RESULTS
Targeting of Phb2 to Mitochondria by a Bipartite Noncleavable Presequence
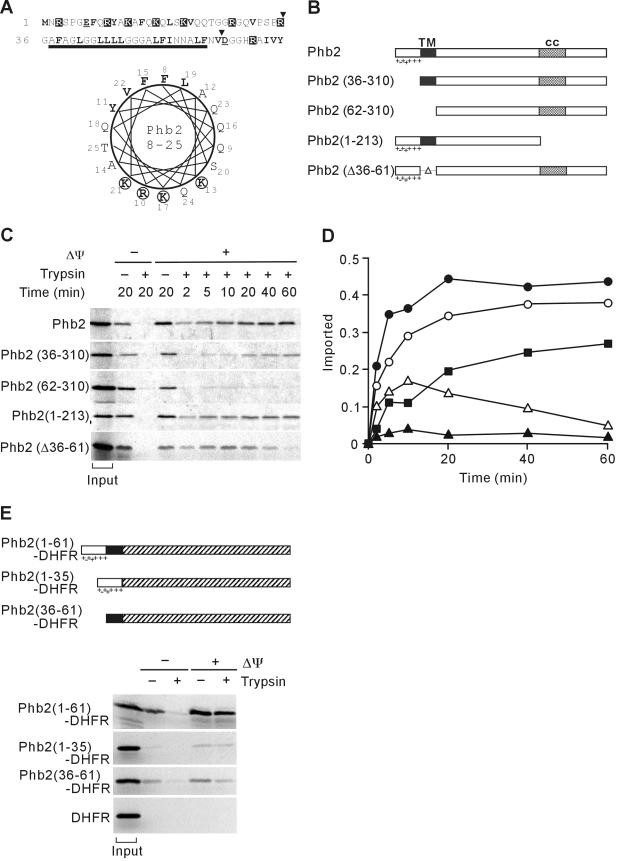
Although generally highly conserved from yeast to human, both Phb1 and Phb2 are more divergent in their N-terminal regions. For Phb2, this segment is composed of a hydrophilic stretch followed by a putative hydrophobic transmembrane domain. Positively charged amino acid residues are enriched within the first 35 amino acid residues, which can form an amphipatic α-helix, a characteristic of mitochondrial sorting sequences (Figure 1A). To examine a potential targeting function of this segment, a series of Phb2 variants were constructed (Figure 1B), synthesized in a cell-free system in the presence of [35S]methionine, and analyzed for their capability to be imported into isolated mitochondria in vitro. Phb2 became resistant to externally added protease in a membrane-potential dependent manner without proteolytic removal of a targeting sequence (Figure 1C). Mitochondrial import, however, was impaired upon deletion of the N-terminal 61 amino acid residues of Phb2 [Phb2(62-310)] demonstrating that this segment harbors essential targeting information (Figure 1, C and D). To dissect the role of hydrophilic and hydrophobic stretches in this segment, we analyzed mitochondrial import of Phb2(36-310) and Phb2(Δ36-61), lacking the positively charged or the putative transmembrane region, respectively. Import of Phb2(36-310) was impaired and occurred with significantly reduced kinetics when compared with Phb2 (Figure 1, C and D). Thus, although the positively charged, N-terminal segment indeed ensures efficient mitochondrial import, additional targeting information is provided by the subsequent hydrophobic segment. Consistently, deletion of the corresponding amino acid residues 36-61 in Phb2 also impaired mitochondrial import (Figure 1, C and D). Notably, Phb2(Δ36-61) did not accumulate within mitochondria, indicating rapid degradation of the newly imported protein.
Figure 1.
Targeting of Phb2 to mitochondria by a noncleavable, bipartite presequence at the N terminus. (A) N-terminal region of Phb2. Amino acid residues 1-70 of S. cerevisiae Phb2 and a helical wheel representation of amino acid residues 8-25 is shown. Charged and hydrophobic residues are highlighted. The bar indicates the predicted transmembrane region of Phb2. The position of N-terminal truncations of Phb2 is marked by arrowheads. (B) Schematic representation of Phb2 variants. TM, predicted transmembrane region; cc, putative coiled-coil region. Charged residues within the N-terminal targeting sequence are indicated. (C) Mitochondrial import of Phb2 and its variants. 35S-labeled precursor proteins were incubated with isolated mitochondria at 16°C for the indicated time in the presence (+Δψ) or absence (-Δψ) of a membrane potential. Nonimported proteins were digested with trypsin when indicated. The quantification of the import kinetics of Phb2 (•), Phb2(36-310) (▪), Phb2(62-310) (▴), Phb2(1-213) (○), Phb2(Δ36-61) (Δ) is shown in D. Total radioactivity incubated with mitochondria is shown as “input” in C and was set to 1 in D. (E) Top, schematic representation of Phb2-DHFR hybrid proteins. Shaded boxes represent the DHFR moiety. Other symbols are as in B. Bottom, import of Phb2-DHFR hybrid proteins into mitochondria. 35S-labeled hybrid proteins were incubated with isolated mitochondria for 20 min at 25°C in the presence (+Δψ) or absence (-Δψ) of a membrane potential. Trypsin was added to digest nonimported proteins when indicated. Mitochondria were analyzed by SDS-PAGE and autoradiography. Signals corresponding to 20% of radiolabeled precursors incubated with mitochondria are shown (input).
To substantiate these findings, we constructed hybrid proteins composed of N-terminal segments of Phb2 and mouse dihydrofolate reductase (DHFR). Phb2(1-61)-DHFR was efficiently imported into mitochondria, demonstrating that the N-terminal 61-amino acid residues are sufficient for mitochondrial targeting (Figure 1E). In contrast, import of Phb2(1-35)-DHFR or Phb2(36-61)-DHFR occurred with low efficiency, indicating that neither the positively charged nor the hydrophobic segment is able to ensure efficient mitochondrial import of a heterologous protein (Figure 1E).
We conclude from these experiments that mitochondrial targeting of Phb2 is ensured by a bipartite noncleavable presequence at the N terminus of Phb2, which is composed of a positively charged and a hydrophobic protein segment.
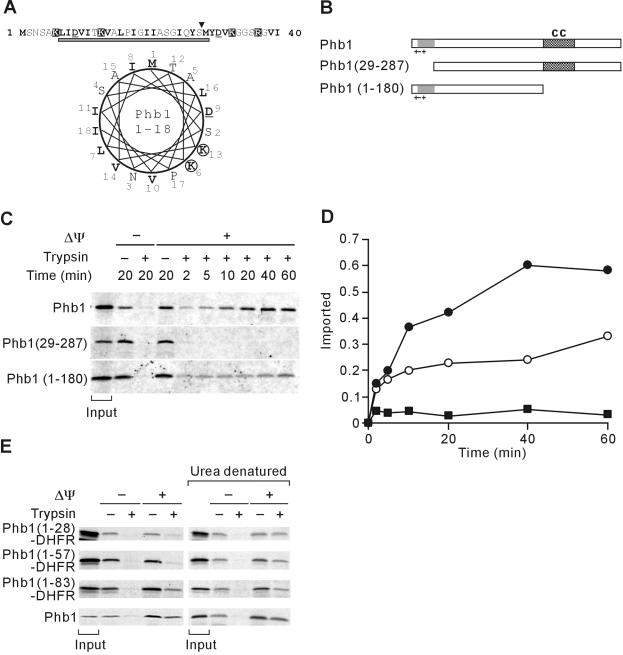
Mitochondrial Targeting of Phb1 by an Unconventional N-Terminal Presequence
Similar to Phb2, Phb1 subunits expose large domains to the intermembrane space and are thought to be anchored N-terminally to the inner membrane. However, neither a positively charged amino acid stretch with a propensity to form a amphipathic α-helix nor a hydrophobic segment predicted to form a membrane-spanning domain are present within the N-terminal region of Phb1 (Figure 2A). Nevertheless, deletion of 28 N-terminal amino acid residues of Phb1 [Phb1(29-287)] abolished import into isolated mitochondria, indicating that the N-terminal segment is indispensable for mitochondrial targeting (Figure 2, C and D).
Figure 2.
Mitochondrial targeting of Phb1 by an unconventional N-terminal presequence. (A) N-terminal region of Phb1. Amino acid residues 1-40 of S. cerevisiae Phb1 and a helical wheel representation of amino acid residues 1-18 is shown and marked as in Figure 1A. The gray bar indicates a potential transmembrane region of Phb1 that is predicted with low scores by using the TMHMM program. (B) Schematic representation of Phb1 and derivatives. Symbols are as in Figure 1B. (C) Mitochondrial import of Phb1 and its variants. 35S-labeled precursor proteins were incubated with isolated mitochondria at 25°C for the indicated time in the presence (+Δψ) or absence (-Δψ) of a membrane potential. Nonimported proteins were digested with trypsin when indicated. The quantification of the import kinetics of Phb1 (•), Phb1(29-288) (▪), and Phb1(1-180) (○) is shown in D. Total radioactivity added to the import reaction is shown as “input” in C and was set to 1 in D. (E) Import of Phb1-DHFR hybrid proteins into mitochondria. 35S-labeled hybrid proteins were incubated with isolated mitochondria for 20 min at 25°C as in C. When indicated, hybrid proteins were precipitated with ammonium sulfate and urea-denatured before import. Signals corresponding to 20% of radiolabeled precursors incubated with mitochondria are shown (input).
When fused to mouse DHFR, however, the 28 N-terminal amino acid residues of Phb1 were not sufficient to promote import of the hybrid protein (Figure 2E). Prolongation of the Phb1-moiety to 57 amino acid residues did not affect targeting to mitochondria, whereas we observed significant import of Phb1(1-83)-DHFR into mitochondria (Figure 2E). To examine a possible impairment of mitochondrial import by folding of the DHFR-moiety, the hybrid proteins also were incubated with isolated mitochondria after denaturation in urea-containing buffer. Under these conditions, Phb1(1-28)DHFR was efficiently imported demonstrating that the first 28 N-terminal amino acid residues contain the complete targeting information (Figure 2E). These experiments demonstrate that Phb1 subunits are targeted to mitochondria by an unconventional noncleavable targeting sequence present at their N terminus.
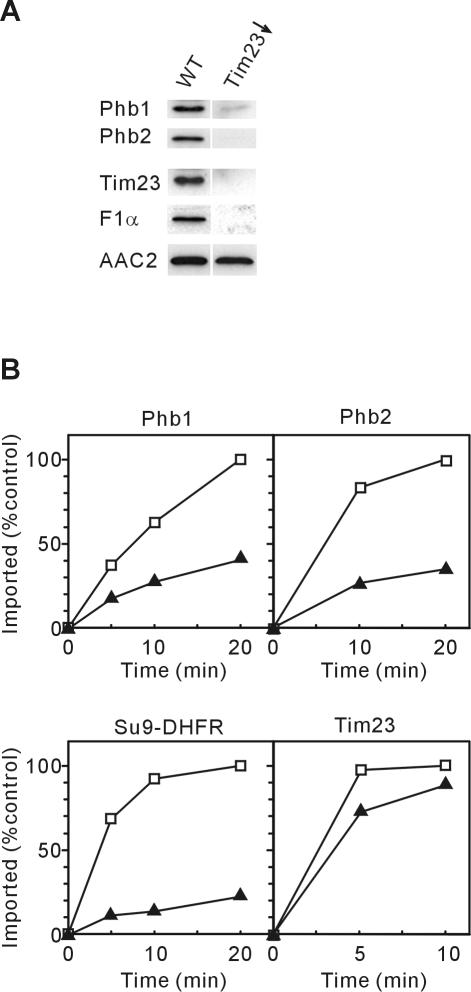
Membrane Insertion Is Mediated by the TIM23-Translocase
The inner membrane harbors two protein translocases for nuclear-encoded preproteins: the TIM23-translocase for presequence-carrying precursor proteins and the TIM22-translocase for polytopic inner membrane proteins (for reviews, see Neupert, 1997; Pfanner and Geissler, 2001). To examine a potential role of the TIM23-translocase for the biogenesis of prohibitins, we used a yeast strain expressing Tim23, a major component of this translocase, from a galactose-inducible promoter. Cells were grown on galactose-free medium for Tim23 depletion and the accumulation of mitochondrial proteins was assessed by immunoblotting. Phb1 and Phb2 were hardly detectable in mitochondria depleted of Tim23, indicating that both proteins require the TIM23-translocase for membrane insertion and stable accumulation within mitochondria (Figure 3A). Similarly, presequence-containing matrix proteins, like the α-subunit of the F1-ATPase, were not detectable in these cells (Figure 3A). In contrast, the carrier protein Aac2, which is imported by the TIM22-translocase (Sirrenberg et al., 1996), accumulated at normal levels in Tim23-depleted mitochondria, demonstrating that the TIM22 pathway was not affected under these conditions (Figure 3A).
Figure 3.
Requirement of the TIM23-translocase for prohibitin biogenesis. (A) Depletion of Tim23 in vivo. Mitochondial membranes were prepared from Tim23(Gal10) cells that were shifted for 24 h to galactose-free medium (Tim23↓). Samples were analyzed by SDS-PAGE and immunodecoration. YPH499 cells were analyzed equally for a control (WT). (B) Import of prohibitins into Tim23-deficient mitochondria. 35S-labeled precursor proteins of Phb1, Phb2, Su9(1-69)-DHFR, and Tim23 were imported into mitochondria isolated from wild-type (mb2) (□) or Tim23(fs) (▴) cells. Protease-resistant, imported protein was quantified by phosphorimaging.
Because Phb1 and Phb2 are functionally interdependent (Berger and Yaffe, 1998; see below), an impaired membrane insertion of either subunit might indirectly affect the accumulation of the other subunit in Tim23-depleted mitochondria. We therefore analyzed the import of radiolabeled Phb1 and Phb2 into mitochondria expressing a C-terminally truncated, mutant variant of Tim23 [Tim23(fs)] (Sirrenberg et al., 1996). Mitochondria isolated from these cells harbored decreased levels of Tim23, whereas Tim22 accumulated at normal levels (Sirrenberg et al., 1996; data not shown). The import of both proteins into these mitochondria was strongly impaired when compared with wild-type mitochondria (Figure 3B). Control experiments demonstrated a general deficiency of TIM23-mediated import, whereas the TIM22-dependent pathway was still functional: the import of the matrix-localized preprotein Su9-DHFR into Tim23(fs) mitochondria was impaired, whereas Tim23, by also using the TIM22-translocase (Leuenberger et al., 1999), accumulated with similar efficiencies in Tim23(fs) and wild-type mitochondria (Figure 3B). Depletion of Tim22 from mitochondria, on the other hand, did not affect import of Phb1 or Phb2 (our unpublished data). We conclude from these experiments that both Phb1 and Phb2 are inserted into the inner membrane by the TIM23-translocase.
Formation of an Assembly Intermediate Containing Phb1 and Phb2
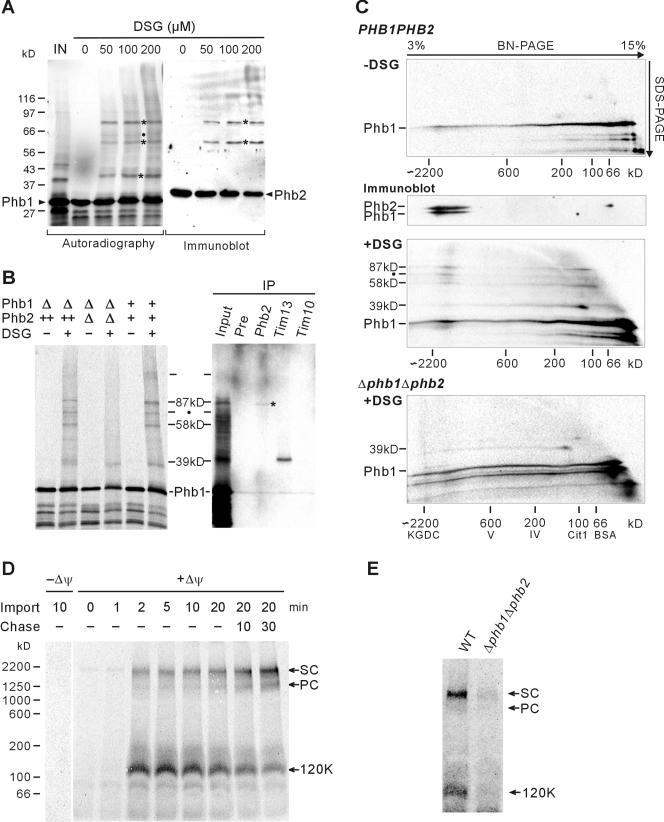
We next analyzed complex formation of Phb1 and Phb2 by chemical cross-linking. 35S-labeled Phb1 was imported into mitochondria that were subsequently incubated with disuccinimidyl glutarate (DSG) (Figure 4A). Phb1-containing cross-link adducts of 39, 58, and 87 kDa accumulated in these mitochondria in a dose-dependent manner (Figure 4A). Several lines of evidence demonstrate an association of newly imported Phb1 with Phb2. First, Phb2-containing complexes of 58 and 87 kDa accumulated upon cross-linking in these mitochondria as revealed by immunoblot analysis by using Phb2-specific antisera (Figure 4A). Second, the formation of these cross-link adducts was found to depend on the presence of Phb2 but not of Phb1, indicating that Phb1-homo-oligomers are not formed. Similar-sized complexes were detected in mitochondria isolated from cells that strongly overexpress and therefore accumulate Phb2, despite the absence of Phb1 (Figure 4B). On the other hand, only the 39-kDa adduct was formed in Δphb1Δphb2 mitochondria (Figure 4B). Third, although with low efficiency, the 87- and 58-kDa cross-link adducts were precipitated with Phb2 antiserum but not with preimmune serum (Figure 4B). We therefore conclude that newly imported Phb1 assembles with Phb2. To examine the formation of large complexes by newly imported Phb1, mitochondria were solubilized after completion of import and chemical cross-linking and extracts were analyzed by two-dimensional blue native/SDS-polyacrylamide gel electrophoresis (BN/SDS-PAGE) (Figure 4C). Whereas preexisting prohibitin complexes were recovered as large protein assemblies (Figure 4C, middle), newly imported Phb1 formed a broad peak, indicating inefficient assembly (Figure 4C). The fractionation was not affected by the presence of excess level of Phb2 within mitochondria (our unpublished data). The cross-link adducts, however, were detected in two distinct complexes with native molecular masses of ∼1-2 MDa and of ∼120 kDa, respectively (Figure 4, C and D). The larger form comigrated with the endogenous prohibitin complex and therefore most likely represents the fully assembled complex.
Figure 4.
Intermediate-sized prohibitin complexes containing Phb1 and Tim13 or Phb1 and Phb2. (A) Chemical cross-linking of newly imported Phb1. After import of 35S-labeled Phb1 and trypsin digestion, mitochondria were subjected to chemical cross-linking by using DSG at the concentrations indicated. Mitochondrial proteins were analyzed by SDS-PAGE and autoradiography (left) or immunodecoration with Phb2-specific antiserum (right). Major cross-link species are indicated by asterisks. A 63-kDa cross-link adduct, which was detected occasionally, is marked with a black dot. The signal corresponding to 50% of radiolabeled precursors incubated with mitochondria is shown (IN). (B) Chemical cross-linking of newly imported Phb1 in mitochondria isolated from wild-type (+/+), Δphb1Δphb2 (Δ/Δ), or Δphb1Δphb2 cells overexpressing Phb2 (-/++). After chemical cross-linking with DSG (150 μM), mitochondria were solubilized and subjected to immunoprecipitation (IP) (right) by using preimmune serum (Pre), Phb2-, Tim13-, and Tim10-specific antiserum. The Phb2-specific antiserum precipitated native Phb2 only very inefficiently (∼1%). A weak cross-link adduct coimmunoprecipitated with Phb2-specific antisera is indicated by an asterisk. (C) BN/SDS-PAGE analysis of cross-link adducts containing newly imported Phb1. After import of Phb1 into mitochondria isolated from YTT163 cells (PHB1PHB2, top) or YGS507 cells (Δphb1Δphb2, bottom), mitochondria were subjected to chemical cross-linking with DSG when indicated. Proteins were solubilized with 0.5% DDM (protein/detergent ratio of 1/2.5) and analyzed by two-dimensional BN/SDS-PAGE. α-Ketoglutarate dehydrogenase complex (KGDC, ∼2.2 MDa), monomeric F1FO-ATP synthase (complex V, ∼600 kDa), monomeric COX complex (complex IV, ∼200 kDa), dimeric citrate synthase (Cit1, 100 kDa), and bovine serum albumin (bovine serum albumin, ∼66 kDa) were used for calibration. The endogenous prohibitin complex was detected by immunoblotting by using Phb1- and Phb2-specific antibodies (middle). (D) Transient accumulation of newly imported Phb1 in intermediate-sized complexes. 35S-labeled Phb1 was imported into YTT160 mitochondria for various times in the absence (-Δψ) or presence (+Δψ) of a membrane potential. After import samples were treated with trypsin to remove nonimported precursors and further incubated at 25°C when indicated (Chase). Membranes were solubilized with 1.0% digitonin (protein/detergent ratio of 1/5) and analyzed by BN-PAGE and autoradiography. The position of the supercomplex of prohibitin with m-AAA protease (2 MDa, SC), prohibitin complex (1.2 MDa, PC), and ∼120-kDa intermediate (120K) are indicated. Dimeric F1FO-ATP synthase (∼1,250 kDa) and the supercomplex composed of bc1 and COX complexes (III2IV2, ∼1000 kDa) were used as additional molecular mass markers. (E) Impaired formation of prohibitin complexes in Δphb1Δphb2 mitochondria. 35S-labeled Phb1 was imported for 20 min into mitochondria isolated from wild-type (WT) or YGS507 cells (Δphb1Δphb2). Samples were processed as in D.
Strikingly, the 39-kDa cross-link adduct was detected in the ∼120-kDa intermediate-sized complex, indicating a transient interaction of a small mitochondrial protein with newly imported Phb1 (Figure 4C). Several low-molecular-weight Tim proteins have been linked to the import of nuclear-encoded mitochondrial proteins (Koehler, 2004). Whereas the Tim9/10 complex is involved in the import of polytopic inner membrane proteins, the Tim8/13 complex plays a role for insertion of some proteins into both the inner or outer membrane (Davis et al., 2000; Paschen et al., 2000; Curran et al., 2002; Vasiljev et al., 2004). To examine a potential involvement of either complex in the biogenesis of prohibitins, coimmunoprecipitation experiments were performed after import of Phb1 into mitochondria and chemical cross-linking. Whereas no cross-link adducts were precipitated with Tim10-specific antiserum, the 39-kDa form was efficiently precipitated with Tim13-specific antiserum, indicating an interaction of Phb1 with the Tim8/13 complex upon import (Figure 4B). Notably, binding of Phb1 to Tim13 did not depend on the presence of Phb2 within mitochondria and therefore seems to occur before the formation of the Phb1/Phb2 assembly intermediate (Figure 4, B and C, bottom).
The interaction of Phb1 with the Tim8/13 complex, however, is not essential for the biogenesis of the fully assembled prohibitin complex. Phb1 and Phb2 accumulated in Δtim8Δtim13 mitochondria at similar levels as in wild-type mitochondria (our unpublished data). Moreover, import of Phb1 occurred with similar efficiencies in wild-type and Δtim8Δtim13 mitochondria and was not affected by lowering the membrane potential across the inner membrane (our unpublished data).
The cross-linking experiments indicate that two ∼120-kDa complexes are formed upon import of Phb1: a Phb1/Tim13 complex and a Phb1/Phb2 complex that may represent an assembly intermediate. To substantiate this conclusion, we analyzed the time course of its accumulation within mitochondria by BN/PAGE (Figure 4D). Radiolabeled Phb1 was imported into wild-type mitochondria that were further incubated after completion of import to allow a more efficient formation of the prohibitin complex. Assembly of newly imported Phb1 was assessed by BN/PAGE (Figure 4D). Whereas 120-kDa forms vanished over time, we observed the formation of the assembled prohibitin complex upon prolonged incubation of mitochondria (Figure 4D). As membranes were solubilized under mild conditions by using digitonin in these experiments, the supercomplex of prohibitins with the m-AAA protease was detected on the BN/PAGE (Figure 4D). The formation of Phb1-containing complexes depended on the membrane potential across the inner membrane and thus seems to require membrane insertion. The 120-kDa form was not detectable after import of Phb1 into Δphb1Δphb2 mitochondria, indicating that these forms represent mainly Phb1/Phb2 complexes (Figure 4E). Consistently, their formation was only slightly affected in mitochondria isolated from Δtim8Δtim13 cells (our unpublished data). These experiments demonstrate the transient accumulation of 120-kDa Phb1/Phb2 complexes upon import of Phb1 into mitochondria and suggest that this form represents an assembly intermediate.
Assembly of the Prohibitin Complex Depends on Coiled-Coil Regions in Phb1 and Phb2
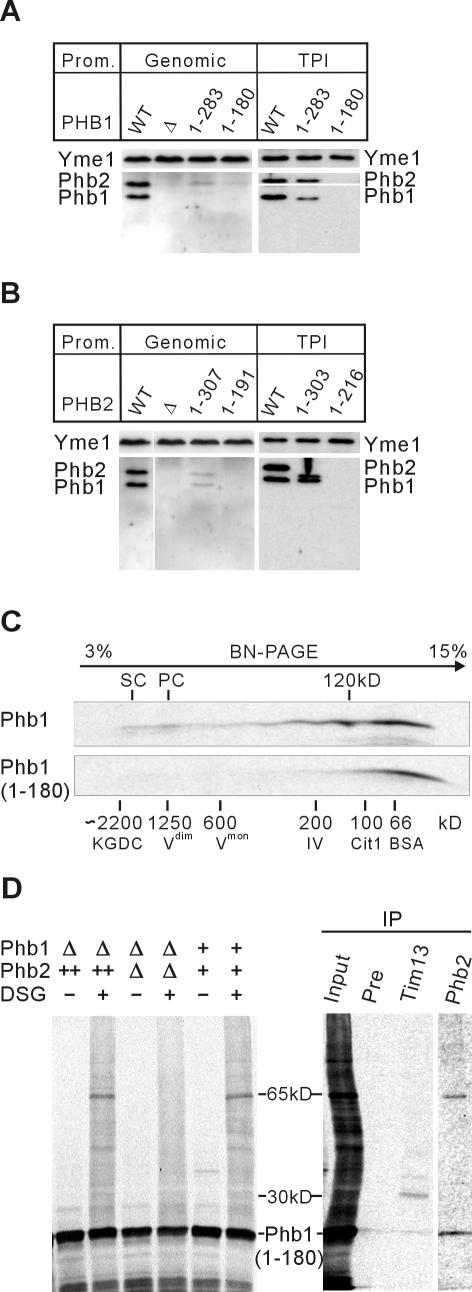
The only conserved sequence motif detectable in prohibitin subunits is a predicted coiled-coil region in the C-terminal part of the polypeptides (amino acid residues 180-224 of Phb1 and 212-253 of Phb2) whose function is presently not understood. The C-terminally truncated variants Phb1(1-180) and Phb2(1-213) were efficiently imported into isolated mitochondria, demonstrating that C-terminal parts of both subunits are dispensable for mitochondrial targeting and transport (Figures 1, C and D, and 2, C and D). Notably, although occurring with initial rates similar to Phb1, import of Phb1(1-180) ceased upon prolonged incubation times (Figure 2D). This might be explained by a decreased solubility of Phb1(1-180) before import or reflect proteolysis of newly imported Phb1(1-180) within mitochondria.
Phb1 and Phb2 are interdependent and accumulate stably only in an assembled state, e.g., nonassembled subunits are rapidly degraded within mitochondria (Berger and Yaffe, 1998). We therefore examined a potential role of the C-terminal coiled-coil regions in both subunits for the assembly of the prohibitin complex. Termination codons were genomically inserted in the PHB1 and PHB2 genes to allow the expression of C-terminally truncated Phb1(1-180) or Phb2(1-191) lacking the coiled-coil regions. In agreement with an impaired assembly of the prohibitin complex, Phb1 or Phb2 did not accumulate in cells expressing Phb2(1-191) or Phb1(1-180) (Figure 5, A and B). We also did not observe a stabilization of Phb1 upon modest overexpression of Phb2(1-216) in Δphb2 cells, nor of Phb2 in the presence of increased levels of Phb1(1-180) in Δphb1 cells (Figure 5, A and B). These results point to an essential function of C-terminal coiled-coil regions in Phb1 and Phb2 for the assembly of the prohibitin complex.
Figure 5.
Assembly of the prohibitin complex depends on C-terminal regions of Phb1 and Phb2. (A and B) Mitochondrial membranes were analyzed by SDS-PAGE and immunoblotting by using Phb1- and Phb2-specific, and as a loading control, Yme1-specific antisera in the following yeast strains. (A) Wild-type cells (WT), Δphb1 cells (Δ), cells expressing C-terminally truncated Phb1(1-180)(YTT216), or Phb1(1-283)(YTT214) from the endogenous promoter (Genomic) and Δphb1 cells expressing Phb1 or the truncated Phb1 variants under the control of a constitutive TPI promoter (pTT46, pTT51, pTT50). (B) Wild-type cells (WT), Δphb2 cells (Δ), cells expressing Phb2 (pTT44), C-terminally truncated Phb2(1-191)(YTT222), or Phb2(1-308)(YTT220) from the endogenous promoter (Genomic) and Δphb2 cells expressing the truncated Phb2 variants Phb2(1-216)(pTT55) or Phb2(1-303)(pTT54) under the control of a constitutive TPI promoter. (C) BN/SDS-PAGE of newly imported Phb1 and Phb1 (1-180). 35S-labeled Phb1 and Phb1(1-180) were imported for 20 min at 25°C into mitochondria isolated from YTT163 cells. Mitochondrial membranes were solubilized by digitonin and subsequently analyzed by BN/SDS-PAGE. PC, prohibitin complex; SC, supercomplex of prohibitins with m-AAA protease. (D) Radiolabeled Phb1(1-180) was imported into mitochondria isolated from wild-type (+/+), Δphb1Δphb2 (Δ/Δ), or Δphb1Δphb2 strains expressing Phb2 (-/++) as in C followed by chemical cross-linking with DSG. When indicated (IP), mitochondrial extracts were analyzed by immunoprecipitation by using Phb2- and Tim13-specific antiserum or preimmune serum (Pre). The lane showing the immunoprecipitate with Phb2-specific antibodies was exposed three times as long as the other lanes.
Surprisingly, deletion of only four or three C-terminal amino acid residues from genomically encoded Phb1 or Phb2, respectively, caused already a destabilization of the respective binding partner (Figure 5, A and B), indicating that the integrity of the C-terminal end of both prohibitin subunits is important for complex formation. Consistently, heaxahistidine peptides fused to the C terminus of Phb1 or Phb2 abolished the assembly of the prohibitin complex (our unpublished data). The impaired complex formation after deletion of short C-terminal segments of Phb1 or Phb2 could be overcome by modest overexpression of Phb2(1-303) or Phb1(1-283), demonstrating that, in contrast to the coiled-coil regions, C-terminal amino acid residues of Phb1 and Phb2 are crucial but not essential for the assembly of the prohibitin complex.
The Coiled-Coil Region of Phb1 Is Required for the Formation of Large Complexes but Dispensable for the Interaction with Phb2 and Tim13
The detection of Phb1/Phb2 assembly by chemical cross-linking and BN/PAGE analyses allowed us to further define the role of the coiled-coil region of Phb1 during the formation of the prohibitin complex. 35S-labeled Phb1 and Phb1(1-180) lacking the C-terminal coiled-coil domain were imported into isolated mitochondria. After completion of import and solubilization of mitochondrial membranes with digitonin, the assembly of the newly imported proteins was assessed by BN/SDS-PAGE (Figure 5C). As observed previously (Figure 4C), Phb1 was detected as a broad peak after electrophoretic fractionation of mitochondrial extracts. It should be noted, however, that small portions of newly imported Phb1 were enriched in intermediate-sized complexes and in a large complex, indicating assembly of the ring complex (Figure 5A). In contrast, assemblies containing Phb1(1-180) were not detected (Figure 5A). These findings are in agreement with the in vivo analysis and indicate that the coiled-coil region of Phb1 is essential for the formation of the prohibitin complex.
Chemical cross-linking was used to examine the requirement of the coiled-coil region of Phb1 for binding to Phb2. Newly imported Phb1(1-180) formed a cross-link of ∼65 kDa (Figure 5D). This adduct was dependent on the presence of Phb2 within mitochondria, and although with low efficiency, could be precipitated with Phb2-specific antibodies (Figure 5D). In addition, we observed the formation of a ∼30-kDa cross-link containing Phb1(1-180), which was precipitated with Tim13-specific antibodies (Figure 5D). We conclude from these experiments that the C-terminal coiled-coil region of Phb1 is dispensable for Tim13-binding and the initial interaction with Phb2, whereas it is required for the formation of the large prohibitin complex.
Prohibitins Form Large Ring-shaped Complexes in the Mitochondrial Inner Membrane
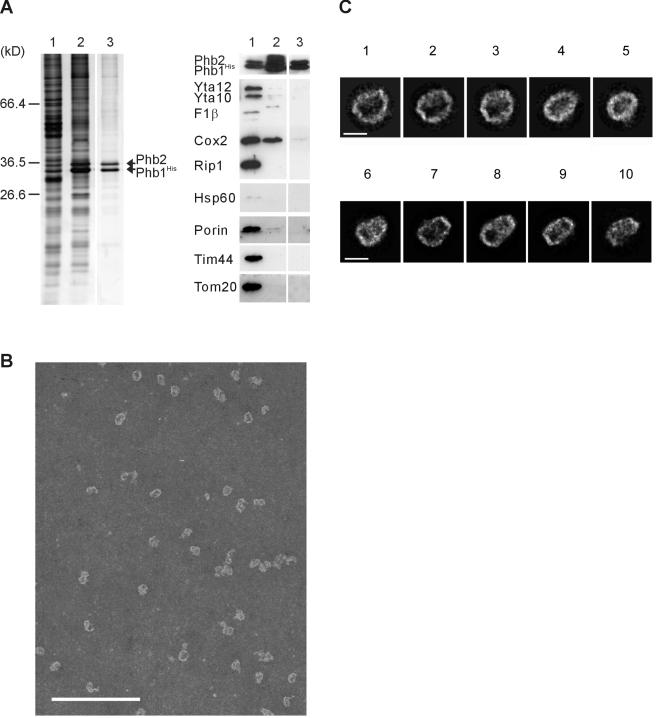
The native molecular mass of the assembled prohibitin complex allows to characterize its molecular architecture by electron microscopy. We therefore purified the prohibitin complex from yeast pursuing the following strategies: First, both Phb1 and Phb2 were expressed simultaneously from one multicopy plasmid under the control of galactose-inducible promoters. This resulted in a ∼20-fold overexpression of Phb1His and Phb2 compared with wild-type cells (our unpublished data). Second, a hexahistidine peptide was fused to the N terminus of Phb1 (Phb1His) to allow affinity purification by metal chelating chromatography. Expression of Phb1His suppressed the growth defect of a PHB1 deletion in Δyta10 cells, demonstrating the functional activity of Phb1His in vivo (our unpublished data). Notably, overexpressed Phb1His and Phb2 were exclusively detected in mitochondria and not mislocalized to other cellular compartments. The amino-terminal hexahistidine tag did not interfere with the import of Phb1His into mitochondria (our unpublished data). Therefore, isolated mitochondria were used as a starting material for purification. Third, mitochondrial membranes were solubilized in DDM, conditions that destabilize the supercomplex of prohibitins and the m-AAA protease. The prohibitin complex was then purified to homogeneity by metal chelating chromatography and glycerol gradient sedimentation (Figure 6A). Abundant mitochondrial proteins, such as Hsp60, or other membrane-bound protein complexes, such as the F1FO-ATP synthase, bc1 complex, TIM or TOM translocases, m-AAA proteases, or porin were not detected by Western blot analysis in the purified fractions (Figure 6A). Similarly, subunits of the 20S proteasome were immunologically not detectable in the purified protein fraction (our unpublished data).
Figure 6.
Single particle electron microscopic analysis of the prohibitin complex. (A) Purification of prohibitin complexes from Δphb1 mitochondria harboring overexpressed Phb1His and Phb2 by metal chelating chromatography and glycerol gradient centrifugation as described in Materials and Methods. Samples were analyzed by SDS-PAGE and silver staining (left) or immunodetection against mitochondrial proteins as indicated (right). Prohibitin-containing fractions are shown. Lane 1, mitochondrial membranes (2 μg [silver staining] or 10 μg [immunodetection]); lane 2, eluate of Ni-NTA chromatography (1/200 or 1/40 of total); lane 3, prohibitin-containing fraction after glycerol gradient centrifugation (corresponding to a glycerol concentration of 24-26%; 1/40 or 1/8 of total). (B) Representative section of a micrograph showing a glycerol gradient fraction enriched in prohibitin complexes stained with 1% phosphotungstate. Bar, 200 nm. (C) Class averages of typical end-on or side-on projection views from prohibitin complexes. The data set was split in two subsets according to either elliptical ring-like (32%) or more rectangular views (68%). Percentages of particles of corresponding average images 1-5 are given as follows: for subset I (top), set to 100% class 1 counts 9%, class 2 6%, class 3 16%, class 4 12%, and class 5 11% of particles; for subset II (bottom), set to 100%, class 6 counts 17%, class 7 17%, class 8 8%, class 9 13% and class 10 8% of particles. Bar, 200 Å.
The purified prohibitin complex was investigated by single particle electron microscopy. The mild purification scheme allowed us to detect the majority of particles as single and stable complexes (Figure 6B). Electron micrographs of negatively stained preparations showed various projection views (Figure 6C). Predominant views included roughly elliptical rings with outer dimensions of 270 × 200 Å and a 160 × 90 Å central stain-filled cavity (Figure 6C, classes 1-5) and more rectangular structures frequently composed of two slightly different halves with overall average dimensions of ∼260 × 170 Å (Figure 6C, classes 6-10). Similar particles were not detected in control experiments with corresponding fractions from Δphb1Δphb2 cells. Based on these observations, we propose that prohibitins form large ring-like complexes on the inner mitochondrial membrane to which they are anchored by hydrophobic N-terminal helices.
DISCUSSION
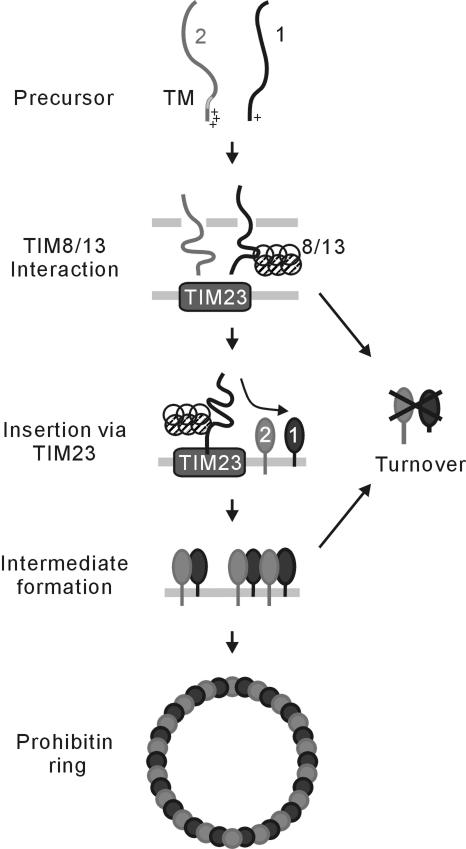
We show that prohibitins form large ring-shaped complexes on the inner membrane of mitochondria and characterize several distinct steps during the biogenesis of this ring structure (Figure 7). 1) Both Phb1 and Phb2 are targeted to mitochondria by unconventional sorting sequences present at their amino terminal regions. 2) Newly imported Phb1 was found in association with Tim13. 3) Phb1 and Phb2 subunits insert into the inner membrane via the Tim23-translocase, where they form assembly intermediates with an apparent molecular mass of ∼120 kDa. 4) These intermediates are proposed to assemble into large ring complexes on the inner membrane, a process dependent on conserved coiled-coil regions present in both Phb1 and Phb2.
Figure 7.
Model for the biogenesis of the ring-shaped prohibitin complex in mitochondria. See Discussion for details. The average stoichiometry of assembly intermediates and the ring complex is speculative. 1, Phb1; 2, Phb2; 8/13, TIM8/13; TM, transmembrane region.
Prohibitin subunits harbor targeting signals within their amino terminal segments that are distinct from that of other intermembrane space proteins whose intramitochondrial sorting has been analyzed (for reviews, see Neupert, 1997; Pfanner and Geissler, 2001). Phb2 subunits carry a bipartite presequence composed of a charged region with the propensity to form an amphipathic helix and a hydrophobic stretch. This is reminiscent of cytochrome c peroxidase and cytochrome b2, soluble proteins in the intermembrane space, as well as cytochrome c1, a tail-anchored protein exposing its catalytic domain to the outer surface of the inner membrane (Hartl et al., 1987; Kaput et al., 1989). Whereas the bipartite presequences of these preproteins are cleaved off in a stepwise manner by specific processing peptidases in mitochondria, Phb2 subunits are not processed upon import. Moreover, the hydrophobic stretch of Phb2 not only harbors sorting information but also serves as a membrane anchor for Phb2 subunits. It therefore has a function similar to transmembrane segments of other inner membrane proteins that harbor one membrane-spanning domain and whose amino terminal segments are targeted to the matrix by a positively charged sorting signal. In contrast to Phb2, the amino terminal region of Phb1 subunits lacks any characteristics of known mitochondrial targeting signals but nevertheless is essential and sufficient for mitochondrial import. A hexahistidine peptide fused to the amino terminus of Phb1 did not interfere with the import of precursors into mitochondria in vivo nor in vitro, indicating that overall positive charge rather than the structure of the amino terminal region is important for the targeting of Phb1. Notably, basic features of the amino terminal segment of Phb1, in particular the distribution of charged amino acid residues, are conserved during evolution. The corresponding region of the rat orthologue of Phb1 can form an amphipatic helix (Ikonen et al., 1995). Our findings therefore set the stage for the analysis of the intracellular sorting of mammalian Phb1 that has been localized not only to the mitochondrial inner membrane but also to the nucleus (Thompson et al., 2001; Wang et al., 2002; Fusaro et al., 2003; Kurtev et al., 2004; Sun et al., 2004).
Chemical cross-linking experiments revealed an association of newly imported Phb1 with Tim13. Tim13 proteins form, with Tim8 proteins a soluble, 70-kDa complex in the intermembrane space that functions in the biogenesis of inner and outer membrane proteins (Leuenberger et al., 1999; Davis et al., 2000; Paschen et al., 2000; Curran et al., 2002; Hoppins and Nargang, 2004). Although we did not observe chemical cross-linking to Tim8, the apparent native molecular mass of the Tim13/Phb1 complex points to binding of Phb1 to the assembled Tim8/13 complex. The association of Phb1 with Tim13 does not depend on Phb2 nor on the C-terminal coiled-coil region of Phb1, suggesting that it occurs to N-terminal regions of Phb1 during early stages of import and before the assembly with Phb2. Although not essential for import, the association with the Tim8/13 complex may trap newly imported Phb1 in the intermembrane space and facilitate its transfer to the inner membrane. Membrane insertion of prohibitins depends on the membrane potential and is mediated by the Tim23-translocase.
Newly imported Phb1 assembles with Phb2 subunits into a subcomplex of ∼120 kDa, which most likely represents a membrane-bound intermediate during the assembly of the prohibitin complex. The formation of this intermediate does not depend on the Tim8/13 complex nor is its apparent native molecular mass altered in Δtim8Δtim13 mitochondria, suggesting that Tim13 is not part of this structure. Vice versa, the native molecular mass of the Phb1/Tim13 complex does not depend on the presence of Phb2, suggesting that two similar-sized but independent complexes are formed upon import of Phb1. It should be noted that the Phb1/Tim13 complex was only detected after cross-linking but could not be detected by BN-PAGE analysis. We propose that intermediate-sized complexes built up of Phb1 and Phb2 oligomerize in the inner membrane and assemble into ring-shaped structures. Coiled-coil regions present in C-terminal regions of Phb1 and Phb2 subunits are crucial for this assembly process but not for the formation of intermediate-sized complexes detected by chemical cross-linking. It should be noted, however, that Phb2 subunits are degraded in yeast cells expressing C-terminally truncated Phb1. These findings indicate that the intermediate complexes reflect labile structures that do not stabilize Phb2 subunits against degradation. We therefore conclude that the formation of stable assemblies of Phb1 and Phb2 depends on coiled-coil regions present in both prohibitin subunits.
Our experiments did not provide any evidence for homodimers of Phb1 and are therefore in agreement with previous chemical cross-linking experiments and structural models predicting a ring-like assembly of alternating Phb1 and Phb2 subunits (Back et al., 2002). Direct support for this model was obtained by single particle electron microscopic analysis of purified prohibitin complexes that revealed large ring-shaped structures with outer dimensions of ∼270 × 200 Å. Given a native molecular mass of the assembled complex of ∼1.0-1.4 MDa (without detergent and lipid), the ring should be composed of ∼16-20 subunits of both Phb1 (31 kDa) and Phb2 (35 kDa). The apparent variability of the shape and size of prohibitin rings might reflect the dynamic nature of the complex that is further indicated by the incorporation of newly imported Phb1 and Phb2 subunits into existing complexes. It is conceivable that different sizes of prohibitin rings differ in the number of subunits, as sterical restraints may be small due to the large number of prohibitin subunits in the ring. The observed irregularities in ring-shaped classes of particles also may arise from imperfect orientation or distortions induced by the preparation or may reflect different conformations of the complexes and thereby be of functional relevance. Therefore, a more extensive electron microscopic analysis has to be awaited to reveal the stoichiometry and detailed architecture of the prohibitin ring complex.
The formation of ring-like complexes sheds some light on potential functions of prohibitins in the mitochondria. Based on the observed interaction of prohibitins with newly synthesized mitochondrial translation products and a limited sequence similarity to chaperonins of the GroEL/Hsp60- class, a chaperone activity has been proposed for prohibitins that might act as “holdases” during the assembly of the respiratory chain (Nijtmans et al., 2000). Indeed, the ring assembly of prohibitins is reminiscent of chaperonins and self-compartmentalizing proteases that provide a sequestered environment within cylindrical structures. However, the diameter of the prohibitin ring is substantially larger than that of these complexes (∼150 Å for GroEL). Moreover, some stain-excluding density within the ring structure may indicate that the prohibitin complex does not have a continuous inner cavity. In view of its dimensions, a scaffolding function of the prohibitin complex is conceivable, which may ensure the organization and integrity of the inner membrane. This would provide an explanation for the functional interaction of prohibitins with various mitochondrial proteins. Complex formation of prohibitins with the m-AAA protease, which regulates the assembly of various protein complexes in the inner membrane, is of particular relevance in this context (Steglich et al., 1999). The ring structure of the prohibitin complex is also in agreement with the proposed role of prohibitins for the maintenance of mitochondrial morphology. In the absence of prohibitins, aberrant mitochondria accumulate in yeast cells lacking the mitochondrial genome and in body wall muscle cells of C. elegans (Berger and Yaffe, 1998; Piper et al., 2002; Artal-Sanz et al., 2003). Notably, prohibitin ring complexes have a similar diameter as cristae junctions, raising the intriguing possibility that prohibitins affect the ultrastructure of the inner membrane. However, further studies are required to examine whether prohibitins have a direct effect on mitochondrial morphology.
Acknowledgments
We thank S. Paschen, D. Mokranjac, and W. Neupert for yeast strains and D. Tils for expert technical assistance. We are grateful to W. Kühlbrandt for critical reading of the manuscript. This work was supported by grants of the Deutsche Forschungsgemeinschaft, the European Union (6th Framework Programme) and the Center for Molecular Medicine Cologne to T.L.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-09-0807. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0807.
References
- Arlt, H., Tauer, R., Feldmann, H., Neupert, W., and Langer, T. (1996). The YTA10-12-complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85, 875-885. [DOI] [PubMed] [Google Scholar]
- Artal-Sanz, M., Tsang, W. Y., Willems, E. M., Grivell, L. A., Lemire, B. D., van der Spek, H., Nijtmans, L. G., and Sanz, M. A. (2003). The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J. Biol. Chem. 278, 32091-32099. [DOI] [PubMed] [Google Scholar]
- Back, J. W., Sanz, M. A., De Jong, L., De Koning, L. J., Nijtmans, L. G., De Koster, C. G., Grivell, L. A., Van Der Spek, H., and Muijsers, A. O. (2002). A structure for the yeast prohibitin complex: structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Sci. 11, 2471-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, K. H., and Yaffe, M. P. (1998). Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 4043-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner, R., Nebauer, R., Schneiter, R., and Daum, G. (2003). Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine biosynthetic machinery with the prohibitin complex of Saccharomyces cerevisiae. Mol. Biol. Cell 14, 370-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates, P. J., Jamieson, D. J., Smart, K., Prescott, A. R., and Hall, P. A. (1997). The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr. Biol. 7, 607-610. [DOI] [PubMed] [Google Scholar]
- Coates, P. J., Nenutil, R., McGregor, A., Picksley, S. M., Crouch, D. H., Hall, P. A., and Wright, E. G. (2001). Mammalian prohibitin proteins respond to mitochondrial stress and decrease during cellular senescence. Exp. Cell Res. 265, 262-273. [DOI] [PubMed] [Google Scholar]
- Curran, S. P., Leuenberger, D., Schmidt, E., and Koehler, C. M. (2002). The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 158, 1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A. J., Sepuri, N. B., Holder, J., Johnson, A. E., and Jensen, R. E. (2000). Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J. Cell Biol. 150, 1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M., and Leith, A. (1996). SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190-199. [DOI] [PubMed] [Google Scholar]
- Fusaro, G., Dasgupta, P., Rastogi, S., Joshi, B., and Chellappan, S. (2003). Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J. Biol. Chem. 278, 47853-47861. [DOI] [PubMed] [Google Scholar]
- Hartl, F. U., Ostermann, J., Guiard, B., and Neupert, W. (1987). Successive translocation into and out of the mitochondrial matrix: targeting of proteins to the intermembrane space by a bipartite signal peptide. Cell 51, 1027-1037. [DOI] [PubMed] [Google Scholar]
- Hoppins, S. C., and Nargang, F. E. (2004). The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279, 12396-12405. [DOI] [PubMed] [Google Scholar]
- Ikonen, E., Fiedler, K., Parton, R. G., and Simons, K. (1995). Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS Lett. 358, 273-277. [DOI] [PubMed] [Google Scholar]
- Kaput, J., Brandriss, M. C., and Prussak-Wiechowska, T. (1989). In vitro import of cytochrome c peroxidase into the intermembrane space: release of the processed form by intact mitochondria. J. Cell Biol. 109, 101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, C. M. (2004). The small Tim proteins and the twin Cx3C motif. Trends Biochem. Sci. 29, 1-4. [DOI] [PubMed] [Google Scholar]
- Kurtev, V., Margueron, R., Kroboth, K., Ogris, E., Cavailles, V., and Seiser, C. (2004). Transcriptional regulation by the repressor of estrogen receptor activity via recruitment of histone deacetylases. J. Biol. Chem. 279, 24834-24843. [DOI] [PubMed] [Google Scholar]
- Leuenberger, D., Bally, N. A., Schatz, G., and Koehler, C. M. (1999). Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 18, 4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabini, R., Masegosa, I. M., San Martin, M. C., Marco, S., Fernandez, J. J., de la Fraga, L. G., Vaquerizo, C., and Carazo, J. M. (1996). Xmipp: an image processing package for electron microscopy. J. Struct. Biol. 116, 237-240. [DOI] [PubMed] [Google Scholar]
- Marco, S., Chaboyen, M., de la Fraga, L. G., Carazo, J. M., and Carrascosa, J. L. (1996). A variant to the random approximation of the reference-free alignment algorithm. Ultramicroscopy 66, 5-10. [Google Scholar]
- McClung, J. K., King, R. L., Walker, L. S., Danner, D. B., Nuell, M. J., Stewart, C. A., and Dell'Orco, R. T. (1992). Expression of prohibitin, an antiproliferative protein. Exp. Gerontol. 27, 413-417. [DOI] [PubMed] [Google Scholar]
- Montano, M. M., Ekena, K., Delage-Mourroux, R., Chang, W., Martini, P., and Katzenellenbogen, B. S. (1999). An estrogen receptor-selective coregulator that potentiates the effectiveness of antiestrogens and represses the activity of estrogens. Proc. Natl. Acad. Sci. USA 96, 6947-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert, W. (1997). Protein import into mitochondria. Ann. Rev. Biochem. 66, 863-917. [DOI] [PubMed] [Google Scholar]
- Nijtmans, L.G.J., de Jong, L., Sanz, M. A., Coates, P. J., Berden, J. A., Back, J. W., Muijsers, A. O., Van der Speck, H., and Grivell, L. A. (2000). Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 19, 2444-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen, S. A., Rothbauer, U., Kaldi, K., Bauer, M. F., Neupert, W., and Brunner, M. (2000). The role of the TIM8-13 complex in the import of Tim23 into mitochondria. EMBO J. 19, 6392-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner, N., and Geissler, A. (2001). Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell. 2, 339-349. [DOI] [PubMed] [Google Scholar]
- Piper, P. W., Jones, G. W., Bringloe, D., Harris, N., MacLean, M., and Mollapour, M. (2002). The shortened replicative life span of prohibitin mutants of yeast appears to be due to defective mitochondrial segregation in old mother cells. Aging Cell 1, 149-157. [DOI] [PubMed] [Google Scholar]
- Radermacher, M., Ruiz, T., Wieczorek, H., and Gruber, G. (2001). The structure of the V(1)-ATPase determined by three-dimensional electron microscopy of single particles. J. Struct. Biol. 135, 26-37. [DOI] [PubMed] [Google Scholar]
- Schägger, H. (2001). Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65, 231-244. [DOI] [PubMed] [Google Scholar]
- Sirrenberg, C., Bauer, M.F.B., Guiard, B., Neupert, W., and Brunner, M. (1996). Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature 384, 582-585. [DOI] [PubMed] [Google Scholar]
- Steglich, G., Neupert, W., and Langer, T. (1999). Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 19, 3435-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L., Liu, L., Yang, X. J., and Wu, Z. (2004). Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J. Cell Sci. 117, 3021-3029. [DOI] [PubMed] [Google Scholar]
- Terashima, M., Kim, K.-M., Adachi, T., Nielsen, P. J., Reth, M., Köhler, G., and Lamers, M. C. (1994). The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J. 13, 3782-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, W. E., Branch, A., Whittaker, J. A., Lyn, D., Zilberstein, M., Mayo, K. E., and Thomas, K. (2001). Characterization of prohibitin in a newly established rat ovarian granulosa cell line. Endocrinology 142, 4076-4085. [DOI] [PubMed] [Google Scholar]
- Van Heel, M., and Frank, J. (1981). Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy 6, 187-194. [DOI] [PubMed] [Google Scholar]
- Vander Heiden, M. G., Choy, J. S., VanderWeele, D. J., Brace, J. L., Harris, M. H., Bauer, D. E., Prange, B., Kron, S. J., Thompson, C. B., and Rudin, C. M. (2002). Bcl-x(L) complements Saccharomyces cerevisiae genes that facilitate the switch from glycolytic to oxidative metabolism. J. Biol. Chem. 277, 44870-44876. [DOI] [PubMed] [Google Scholar]
- Vasiljev, A., Ahting, U., Nargang, F. E., Go, N. E., Habib, S. J., Kozany, C., Panneels, V., Sinning, I., Prokisch, H., Neupert, W., Nussberger, S., and Rapaport, D. (2004). Reconstituted TOM core complex and Tim9/Tim10 complex of mitochondria are sufficient for translocation of the ADP/ATP carrier across membranes. Mol. Biol. Cell 15, 1445-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Poehlmann, R., and Philippsen, P. (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10, 1793-1808. [DOI] [PubMed] [Google Scholar]
- Wang, S., Fusaro, G., Padmanabhan, J., and Chellappan, S. P. (2002). Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene 21, 8388-8396. [DOI] [PubMed] [Google Scholar]