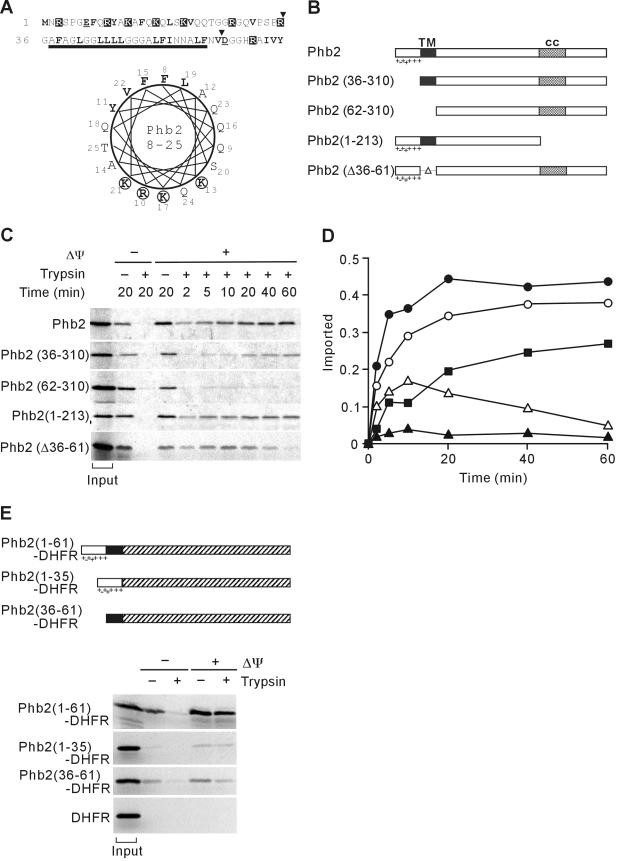
Figure 1.
Targeting of Phb2 to mitochondria by a noncleavable, bipartite presequence at the N terminus. (A) N-terminal region of Phb2. Amino acid residues 1-70 of S. cerevisiae Phb2 and a helical wheel representation of amino acid residues 8-25 is shown. Charged and hydrophobic residues are highlighted. The bar indicates the predicted transmembrane region of Phb2. The position of N-terminal truncations of Phb2 is marked by arrowheads. (B) Schematic representation of Phb2 variants. TM, predicted transmembrane region; cc, putative coiled-coil region. Charged residues within the N-terminal targeting sequence are indicated. (C) Mitochondrial import of Phb2 and its variants. 35S-labeled precursor proteins were incubated with isolated mitochondria at 16°C for the indicated time in the presence (+Δψ) or absence (-Δψ) of a membrane potential. Nonimported proteins were digested with trypsin when indicated. The quantification of the import kinetics of Phb2 (•), Phb2(36-310) (▪), Phb2(62-310) (▴), Phb2(1-213) (○), Phb2(Δ36-61) (Δ) is shown in D. Total radioactivity incubated with mitochondria is shown as “input” in C and was set to 1 in D. (E) Top, schematic representation of Phb2-DHFR hybrid proteins. Shaded boxes represent the DHFR moiety. Other symbols are as in B. Bottom, import of Phb2-DHFR hybrid proteins into mitochondria. 35S-labeled hybrid proteins were incubated with isolated mitochondria for 20 min at 25°C in the presence (+Δψ) or absence (-Δψ) of a membrane potential. Trypsin was added to digest nonimported proteins when indicated. Mitochondria were analyzed by SDS-PAGE and autoradiography. Signals corresponding to 20% of radiolabeled precursors incubated with mitochondria are shown (input).