Figure 4.
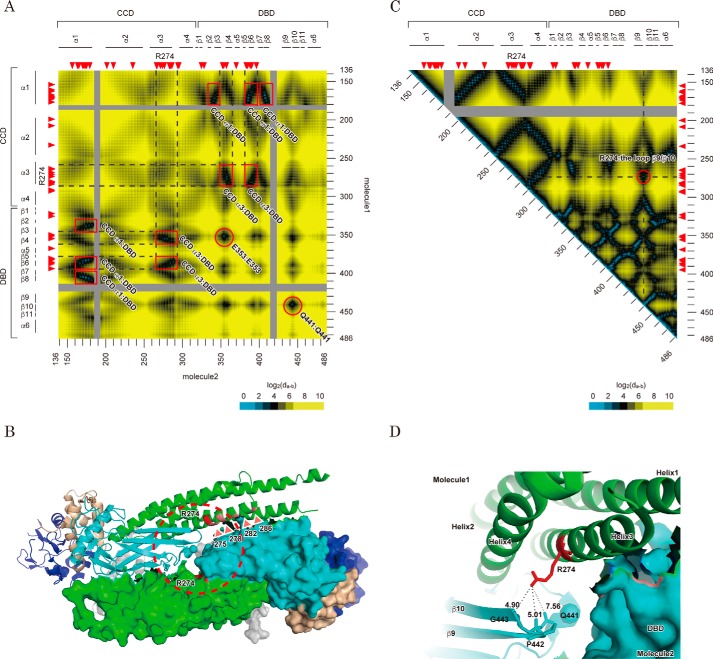
Structural features of Arg-274 in an anti-parallel STAT1 dimer structure. A, interfaces of the anti-parallel conformation of the STAT1 dimer. The heat map depicts the distances from a residue of one component of the anti-parallel STAT1 dimer to that of the other. Possible contact sites of the dimer are marked with a red square or circle. All distances were calculated based on barycentric coordinates of the residues in an anti-parallel dimer structure (Protein Data Bank entry 1YVL). Scale is shown at the bottom. Amino acid number of the residues (right and bottom) and domain and secondary structure of the protein (left and top) are illustrated beside the heat map. Red arrowheads, position of CMC-related GOF mutations. B, locations of the GOF residues characterized in Fig. 1C are illustrated in an anti-parallel STAT1 structure. C, heat map illustrates the distance between two residues of the same STAT1 molecule. All distances were calculated as described in A by using the coordinates of a parallel dimer structure (Protein Data Bank entry 1BF5). A possible attachment site of CCD with DBD is circled in red. D, intramolecular interaction between Arg-274 and Gln-441. The structure surrounded by Arg-274 indicated by a dashed circle in B is enlarged. Each subunit of the dimer is shown as a ribbon diagram and protein surface, respectively. The domains are colored in gray (ND), green (CCD), cyan (DBD and LK), and dark blue (SH2). Distances between the indicated atoms were calculated by the measurement tool packaged in PyMOL.