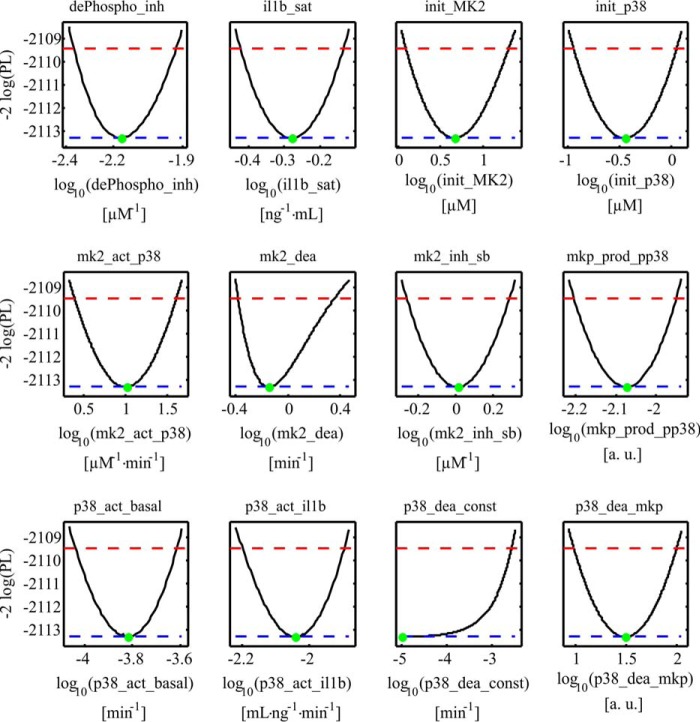
Figure 5.
Parameter likelihood profiles. Likelihood profiles of the dynamic model parameters (dePhospho_inh, il1b_sat, init_MK2, init_p38, mk2_act_p38, mk2_dea, mk2_inh_sb, mkp_prod_pp38, p38_act_basal, p38_act_il1b, p38_dea_const, and p38_dea_mkp). Black lines represent the profile likelihood. The parameter set of the optimum is indicated with a green dot. Red lines indicate the 95% confidence level. Blue lines indicate the global minimum of the parameter estimation process. The analysis revealed that all model parameters are structurally identifiable. The parameter p38_dea_const is practically non-identifiable. The parameter profile suggests that the parameter is compatible with zero; i.e. the constant dephosphorylation rate of p38MAPK is not relevant for our system.