Figure 1.
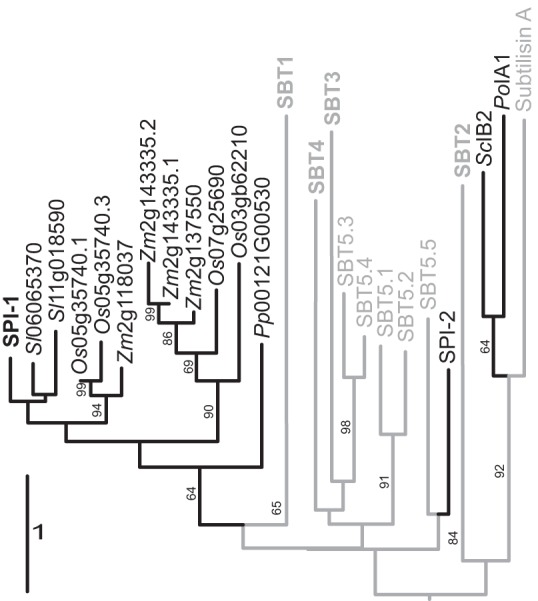
Phylogenetic relationship of propeptides and I9 inhibitors. Maximum likelihood tree was calculated on the Genome-to-Genome Distance Calculator web server and rooted by midpoint rooting of SPI-1 with plant homologs (black) from tomato (Sl); Sl replaces Solyc in tomato gene identifiers, rice (Oryza sativa (Os)), and maize (Zea mays (Zm); Zm replaces GRMZM in maize gene identifiers), also including propeptides of subtilases (gray) from Arabidopsis SBT subfamilies 1–5, subtilisin A from B. licheniformis, and the fungal I9 inhibitors and Arabidopsis SPI-2 (black). The PPs of Arabidopsis SBT subfamilies 1–4 clustered in four distinct clades; these clades were collapsed and are labeled SBT1, -2, -3, and -4, respectively. The branches are scaled in terms of the expected number of substitutions per site. Maximum likelihood bootstrapping values are provided on the left of the branches when larger than 60%.
