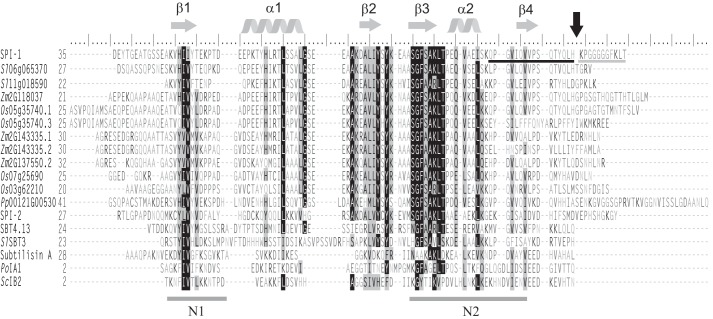
Figure 2.
Amino acid sequence alignment of SPI-1, its plant homologues, SPI-2, the propeptides of AtSBT4.13, SlSBT3, subtilisin A, and two fungal I9 inhibitors after removal of the predicted N-terminal signal peptides (SignalP 3.0; Ref. 64). The sequence alignment with secondary structure prediction was generated using PROMALS3D (33). Highly conserved residues (identical in >70% of the sequences) and partially conserved residues are shaded in black and gray, respectively. Conserved α-helices and β-strands are indicated as gray helices or arrows above the alignment. Conserved hydrophobic regions N1 and N2 are underlined. The C-terminal cleavage site of SPI-1 is marked by a black arrow. ESI-MS- and MALDI-TOF MS-identified peptides flanking the cleavage site are underlined in black and gray, respectively. Residues are numbered from the first Met. Os, O. sativa; Zm, Z. mays.