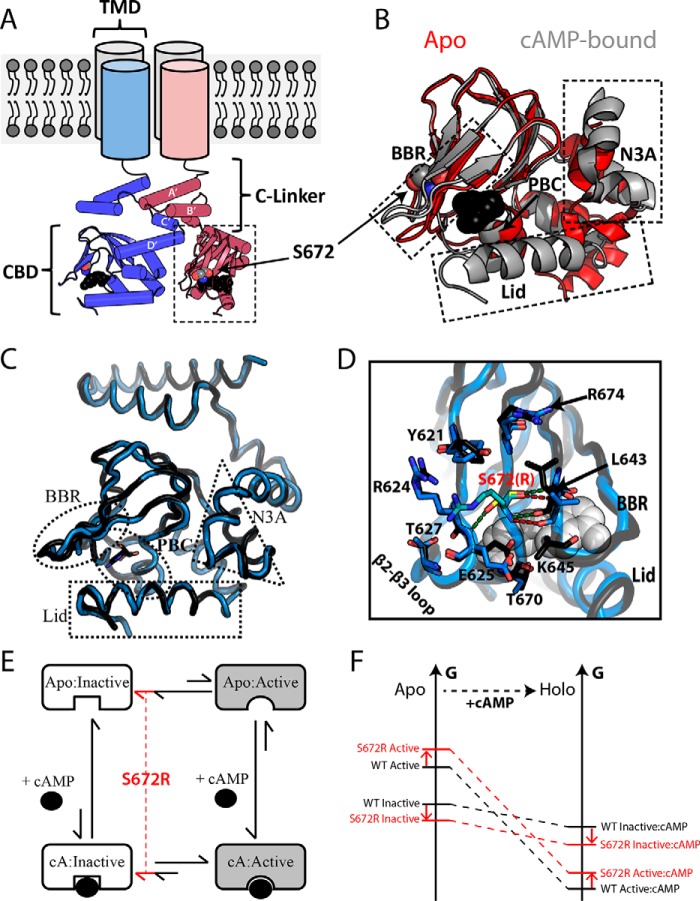
Figure 1.
Structural architecture of the HCN4 intracellular region. A, topology of tetrameric HCN4 channels. The TMD is connected to the CBD through an α-helical domain known as the C-linker, which also serves as the main point of contact between protomers within the gating tetramer. Ser-672 and cAMP are shown as gray and black spheres, respectively. B, overlay of the apo (red; PDB code 2MNG) and cAMP-bound (gray; PDB code 3OTF) CBDs of WT HCN4. Select regions (i.e. N3A, BBR, and Lid) that undergo cAMP-dependent conformational changes are indicated with dashed boxes. cAMP and Ser-672 are depicted as in A. C, structural overlay of holo-WT (black, PDB code 3OTF) and holo-S672R (blue, PDB code 4HBN) HCN4 CBDs. D, changes in key contacts mediated by Ser-672 (black ribbon, yellow sticks) and S672R (blue ribbon, cyan sticks). Polar contacts for Ser-672 and S672R are depicted with green and red dashed lines, respectively. Selected residues that form interactions with S672(R) or that experience side-chain reorientations are shown as sticks. cAMP is shown as white spheres. E, four-state thermodynamic cycle to model allostery for the HCN CBD, in which the auto-inhibitory equilibrium is coupled to the cAMP-binding equilibrium through the active versus inactive state selectivity of cAMP. A hypothetical mechanism for the bradycardia-induced S672R mutant posits that the auto-inhibitory CBD equilibrium is shifted by the mutation further toward the inactive state (red lines). F, simplified free energy diagram illustrating that the hypothesized mutation-induced shift of the auto-inhibitory equilibrium toward the inactive state may arise either from a stabilization of the inactive state and/or from a destabilization of the active state. This scheme assumes that the state-specific association constants are not significantly affected by the mutation.