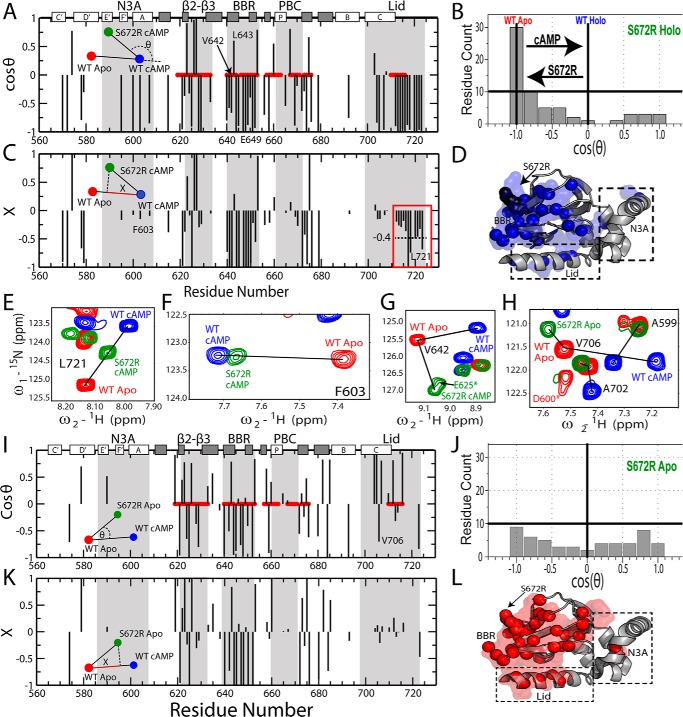
Figure 3.
Chemical shift projection analysis (CHESPA) of the HCN4 S672R mutation. A, cos(θ) values computed as in Equation 3, as explained in the text. Selected residues reported in E–H are labeled. B, distribution of cosθ values from A, revealing a net predominance of negative cos(θ) values. C, fractional activation (X) values for the perturbation vector relative to the activation vector, computed as per Equation 4. The red box highlights a subset of lid residues with markedly negative X values, pointing to a significant shift toward the inactive state. Selected residues reported in E–H are labeled. D, 3D map of CHESPA outliers, defined as residues with either |cosθ| <0.9 or |X| >1.1. Such outliers reveal deviations from the two-state equilibrium, such as those arising from nearest neighbor effects or population of a third state. E, representative NH-HSQC cross-peaks for a residue from the C-terminal lid region with a holo-S672R X value close to −0.4 (C, red box). F, similar to E, but for a representative residue with a holo-S672R X value close to −0.1. G, representative residue (Val-642) from the S672R holo spectra that exhibited a non-linear chemical shift change relative to the WT apo- and holo-states. H, representative residues from the S672R apo spectra compared with WT apo- and holo-states that displayed non-linear chemical shift changes. This non-linearity is also evident from the CHESPA analysis of the apo-S672R HCN4 CBD, shown in I and J, revealing a nearly uniform cos(θ) distribution (I) between the −1 and 1 extremes. Selected residues reported in E–H are labeled in I. K, X values for apo-S672R residues. L, similar to D but for the apo-S672R CHESPA outliers.