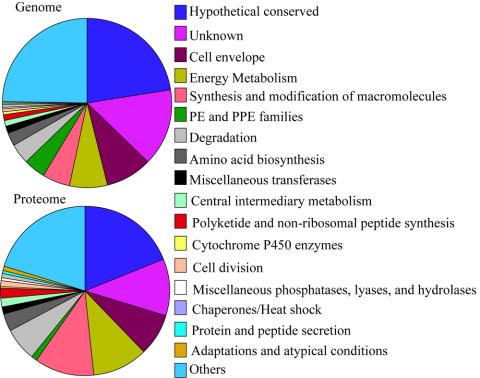
Figure 4.
The plot of variations in functional distribution of genome-encoded proteins compared with those of the proteome. The numerical labels on the pie charts correspond to the functional classes shown on the right-hand side. The sizes of the various pies of the chart show varying percentages of proteins predicted in the genome or identified in our proteome profile. Functional classes listed in Table 1 that did not vary in both the genome and proteome were grouped together and listed under “Others,” which include (I.J) Broad regulatory functions, (IV.B) IS elements, Repeated sequences, and Phage, (III.A) Transport/binding proteins, (I.G) Biosynthesis of cofactors, prosthetic groups and carriers, (II.B) Degradation of macromolecules, (I.H) Lipid Biosynthesis, (I.F) Purines, pyrimidines, nucleosides and nucleotides, (IV.A) Virulence, (III.F) Detoxification, (IV.D) Antibiotic production and resistance, (IV.J) Cyclases, (IV.E) Bacteriocin-like proteins, (IV.G) Coenzyme F420-dependent enzymes, (IV.K) Chelatases, and (I.E) Polyamine synthesis.