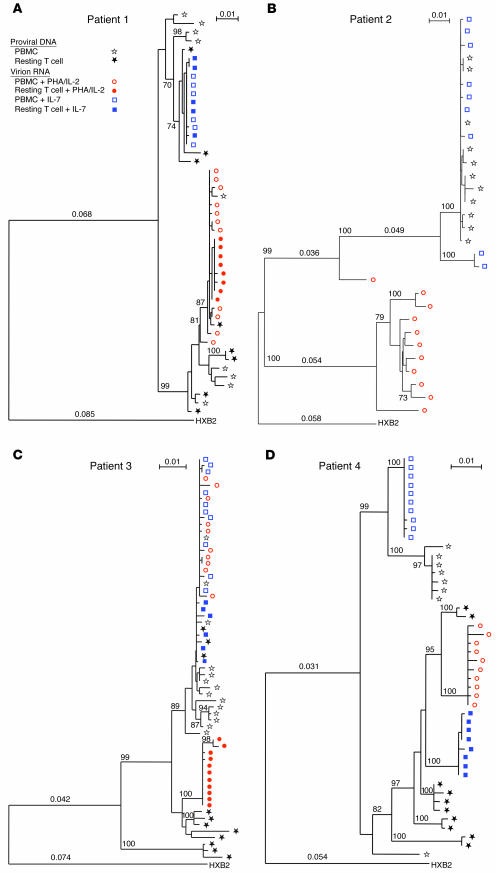
Figure 1.
Phylogenetic analyses of HIV-1 envelope gene sequences of proviral DNA and cytokine-induced virion RNA from the same cell populations. The full-length of gp120 V1 to V5 regions were cloned and sequenced directly from proviral DNA of PBMCs and purified, resting CD4+ T lymphocytes, and IL-7 or PHA/IL-2–induced virion RNA for 4 patients (numbers 1–4, A–D). Proviral DNA sequences are presented by black stars (open stars for those strains from PBMC and filled for those of resting CD4+ T lymphocytes). IL-7–induced viral RNA sequences are indicated by blue squares (open squares for PBMC and IL-7–induced and filled squares for resting CD4+ T lymphocyte and IL–induced). PHA/IL-2–induced viral RNA sequences are indicated by red circles (open circles for those induced by PBMC with PHA/IL-2 and filled circles for those induced by resting CD4+ T- lymphocyte with PHA/IL-2). Growth of patient 4’s virus from PBMCs with PHA/IL-2 was from 10/04/2001. All sequences are shown with HXB2 (GenBank accession no. K03455) as the reference outgroup. Branch lengths are drawn in proportion to the number of nucleotide substitutions per site, and bootstrap probabilities (1,000 iterations) exceeding 70% for each node are noted.