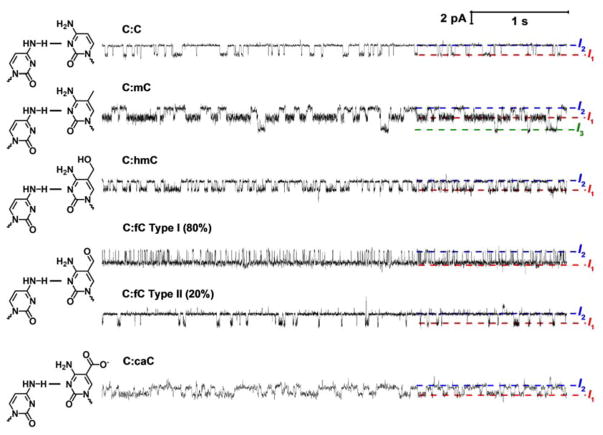
Figure 4.
Substituting the cytosine base at the mismatch site in the shorter (23-mer) strand for methylcytosine (mC) or one of is oxidative derivatives changes the base flipping kinetics. (A) Representative current time traces from a 6 second window of a single DNA capture event demonstrating measurement of base flipping at a C:X mismatch site where X is mC, hmC, fC or caC. Note that two event types (I and II) are observed for fC, with type I comprising 80% of events. This topic is addressed later in the text. Uninterrupted current-time traces demonstrating sequential capture and release of multiple DNA duplexes for each modification are shown in Figures S4 – S7.