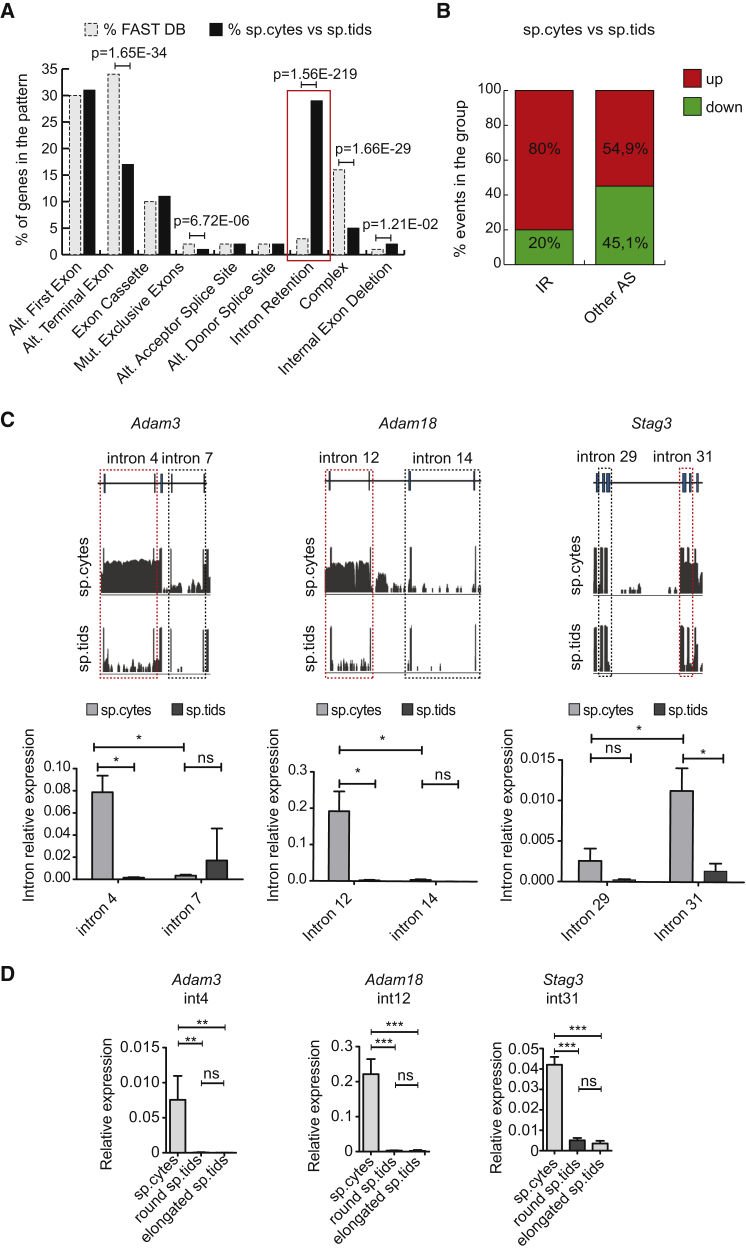
Figure 2.
Intron Retention Is a Key Feature of the Male Meiotic Transcriptome
(A) Bar graph showing percentages of events annotated in FAST-DB (dashed, light gray columns) and of those differentially regulated between spermatocytes (sp.cytes) and spermatids (sp.tids) (black columns) within each AS pattern. p Values above the graph indicate a significant enrichment for regulated events within specific AS pattern in our dataset compared with the reference database (modified Fisher's test). Red box highlights the IR pattern.
(B) Bar graph representing percentages of up- and downregulated events among exonic and intronic splicing events. Enrichment in upregulated events in IR with respect to all other AS events was statistically significant (p = 4.9 × 10−61; χ2 test).
(C) (Top) Visualization of the RNA-seq reads profile of the intron-retaining genes Adam3, Adam18, and Stag3 in sp.cytes (upper graph) and sp.tids (lower graph). Sequence reads (vertical gray lines), exons (blue boxes), and introns (horizontal lines) are shown. (Bottom) Bar graphs showing results of qPCR analyses for the expression of the retained and non-retained introns (red and black dashed boxes, respectively) relative to their flanking exons in sp.cytes and sp.tids (mean ± SD, n = 3; ∗p ≤ 0.05, ns = not significant; one-way ANOVA).
(D) Bar graphs showing results of qPCR analyses for the expression of indicated introns relative to spliced product of their flanking exons in sp.cytes, and round and elongated sp.tids (mean ± SD, n = 4; ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; ns, not significant; one-way ANOVA).
See also Figures S2 and S3.