Figure 3.
IRTs Are Highly Stable mRNAs Retained in the Nucleus of Meiotic Cells
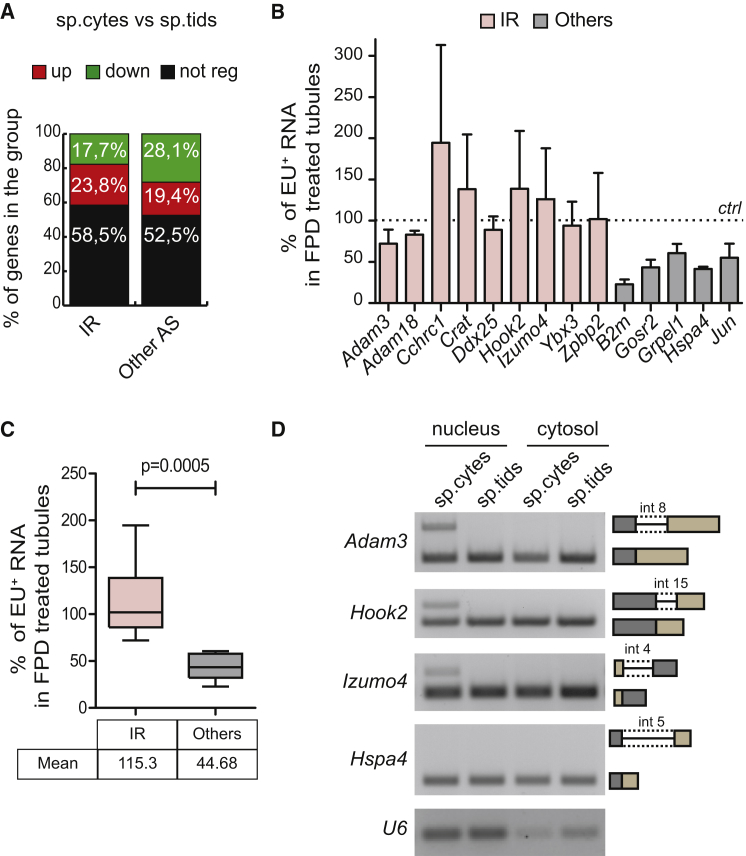
(A) Bar graph showing percentages of expression-regulated genes within the groups of intron-retention and other splicing-regulated genes. A significant association was found between the gene regulation and the type of event (p = 0.0156; χ2 test).
(B) Bar graph showing qPCR analysis using primers spanning exon-intron regions for EU-labeled RNA transcripts of indicated genes isolated from total RNA of seminiferous tubules of 20-dpp (days post-partum) mice treated or not for 1 hr with 1 μM FPD. Results are expressed as percentage of the EU-labeled RNA pulled down relative to control condition, arbitrarily set to 100% (dashed line in the graph, mean ± SD, n = 3).
(C) Box plot showing the distribution of the mean percentages of EU-labeled RNA transcripts pulled down as described in (B) in the IR-regulated genes and other expressed genes. Whiskers indicate 1.5 interquartile range. p Value indicates a significant difference between means of the two groups (Welch's t test).
(D) PCR analysis of the subcellular localization of the IRTs of Adam3, Hook2, and Izumo4 genes. The properly spliced genes Hspa4 and U6 were evaluated as control.
See also Figure S4.