FIGURE 1.
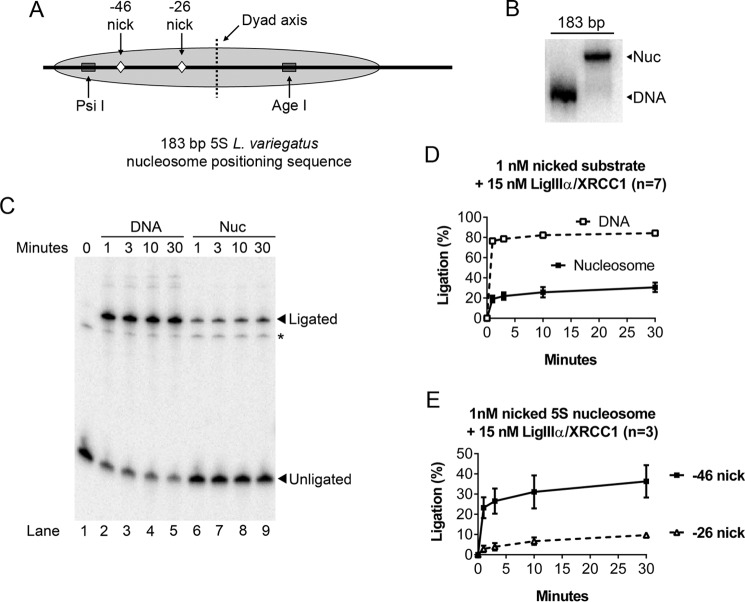
Evidence that spontaneous partial unwrapping of DNA from the histone octamer enables LigIIIα-XRCC1 to discover nicks in nucleosomes. A, schematic depicting features of the nucleosomes used in the experiments shown in Figs. 1–6. The gray oval represents the predominant translational position of the 5S rDNA nucleosome; minor translational variants, in which the octamer is positioned ∼10 bp to either the left or right, also occurred (16, 19, 58) but did not affect the interpretation of the results. Ligatable, single-strand DNA nicks were positioned either −46 or −26 nt away from the nucleosome center (the dyad axis). Also shown are restriction endonuclease cleavage sites used in Fig. 6 to monitor nucleosome integrity during ligation. B, representative nucleosome (Nuc) preparation examined by native PAGE. Nucleosomes were assembled with 2.5 nm labeled, nicked DNA in the presence of 200 nm of nonspecific cold carrier (to ensure nucleosome stability) and then further diluted for subsequent experiments. The presence of this nonspecific DNA had no effect on ligation efficiency (refer to Fig. 4 for competition assays). C, representative 8% denaturing gel used to monitor ligation of nicks in nucleosomes and naked DNA controls as detailed under “Experimental Procedures.” Ligation products migrate as 183-nt bands. The minor band denoted by an asterisk corresponds to unligated DNA located 3′ to the nick and did not interfere with the quantification of ligation products. D, LigIIIα-XRCC1 ligated a significant fraction of the DNA nicks at position −46 even though these nicks faced into the histone octamer, making them accessible only during episodes of spontaneous partial unwrapping of DNA from the histone octamer (see text for further discussion). E, ligation efficiency dropped a further ∼3-fold for nicks positioned 20 nt further in from the nucleosome edge (position −26). This supports the hypothesis that nucleosome unwrapping is rate-limiting LigIIIα-XRCC1. Error bars represent standard deviations.