FIGURE 4.
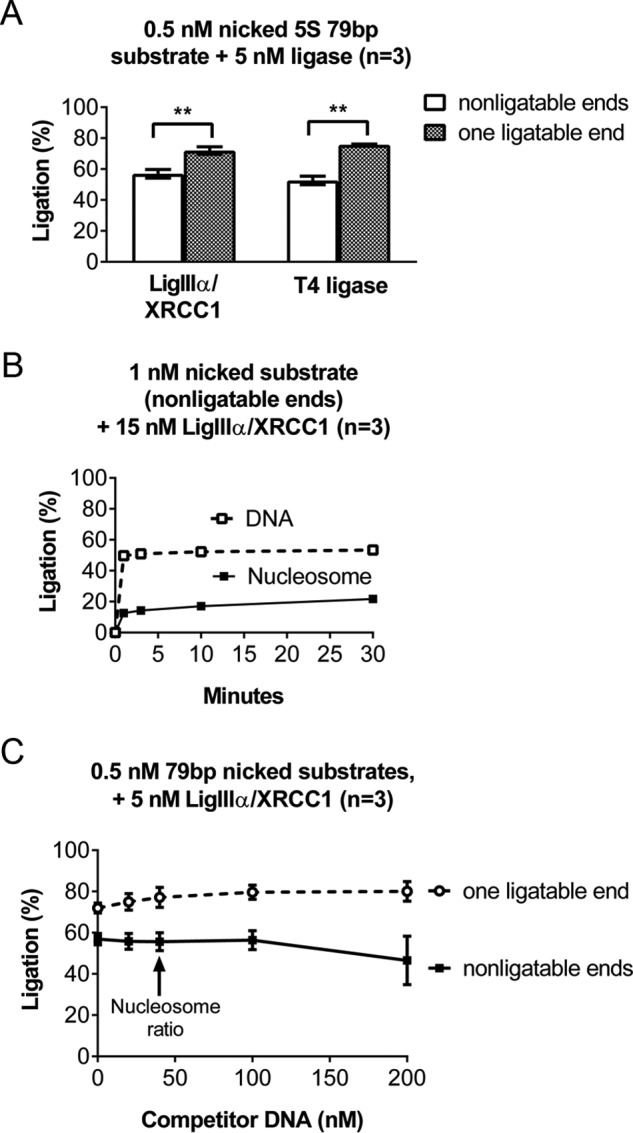
The efficiency with which LigIIIα-XRCC1 discovers nicks in both naked DNA and nucleosomes is enhanced by its capacity to bind DNA ends. A, to examine the impact of DNA end binding by LigIIIα-XRCC1, we prepared a 79-bp DNA fragment containing a single nick and OH groups at both the 5′ and 3′ ends. (The DNA was a truncated version of that used to assemble 5S rDNA-based nucleosomes for many of the experiments reported in this paper.) We labeled this DNA by the addition of 32P to an internal site, producing DNA that is not competent for end binding or blunt end ligation. We also labeled a second aliquot of DNA, identical except for one end containing a 5′-phosphate group. We incubated these differently ended DNAs (0.5 nm) with 5 nm LigIIIα-XRCC1 for 30 min at 37 °C, separated substrate and products on denaturing gels such as those shown in Fig. 1C, and quantified the fraction of substrate ligated using phosphorimaging. The presence of a phosphate group at the 5′ end significantly enhanced ligation. This enhancement was also evident in control reactions using T4 DNA ligase. B, the effect of 5′-phosphate-dependent, DNA end binding on ligation of nicks in nucleosomes. The conditions in these reactions were identical to those in Fig. 1D, except for use of an internally labeled substrate that lacked 5′-phosphate residues and thus would not support DNA end binding. As with the naked DNA substrates, ligation efficiency was significantly reduced (from ∼30% to ∼20%) in the absence of 5′-phosphates. C, to ligation reactions containing the same substrates used in A, we added 0–200 nm naked, unlabeled carrier DNA lacking 5′-phosphate residues. The results indicate that this carrier DNA did not suppress ligation, even at concentrations well above the 80-fold excess carrier used in all other assays. Importantly, we did not detect any dimeric, blunt end ligation products in any of our experiments. Error bars represent standard deviations; ** = p < 0.01.
