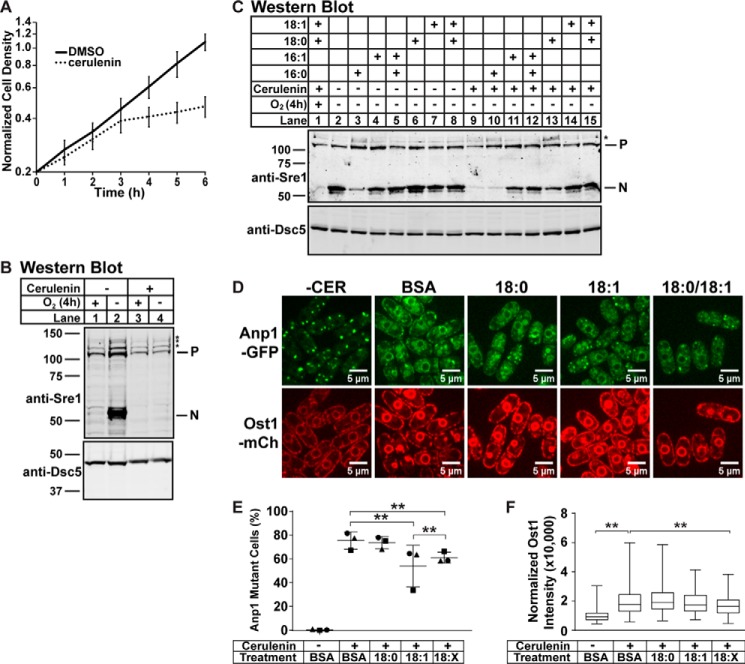
FIGURE 7.
Disrupting fatty acid homeostasis blocks general membrane transport and Sre1 cleavage. A, WT cells were grown in liquid culture in 112 nm CER or 0.1% (v/v) DMSO for 6 h. Cell density was measured by absorbance at 600 nm. Data points are the average of four biological replicates. Error bars are 1 S.D. Absorbance at time 0 among the samples ranged from 0.10 to 0.29. To focus on the difference in growth rate between conditions, data were normalized for each sample to a value of 0.2 at time point 0 before averaging. B and C, Western blots, probed with monoclonal anti-Sre1 IgG (5B4) and polyclonal anti-Dsc5 (for loading) and imaged by LI-COR Biosciences Odyssey CLx, of lysates treated with alkaline phosphatase for 1 h from WT cells. Cells were grown in 112 nm CER or 0.1% (v/v) DMSO for 2 h under normoxia. Cells were then washed, resuspended in fresh medium, and grown without cerulenin for 4 h in the presence or absence of oxygen (B and C) with the indicated BSA-conjugated fatty acid supplemented (C). P and N denote precursor and cleaved N-terminal transcription factor forms, respectively. Asterisks (*) indicate nonspecific bands. Blots are representative of two biological replicates. D, anp1-GFP ost1-mCherry cells were precultured for 1 h in the indicated BSA-conjugated fatty acid. Then 112 nm CER or 0.1% (v/v) DMSO (−CER) was added, and cells were cultured for an additional 3 h before imaging by confocal microscopy. 18:X indicates supplementation of both 18:0 and 18:1. Images are representative of three biological replicates. Scale bar, 5 μm. E, quantification of cells with altered Anp1-GFP localization from D of three biological replicates denoted by different marker symbols. n ≥ 100 cells per replicate. Error bars are mean ± 1 S.D. (**, p < 0.01 by χ2 analysis with Bonferroni correction). E, box-and-whisker quantification of normalized Ost1-mCherry signal intensity from D of n ≥ 300 cells from three biological replicates. Whiskers are the maximum and minimum intensity values (**, p < 0.01 by one-way ANOVA and post hoc multiple comparisons with Bonferroni correction). mCh, mCherry.