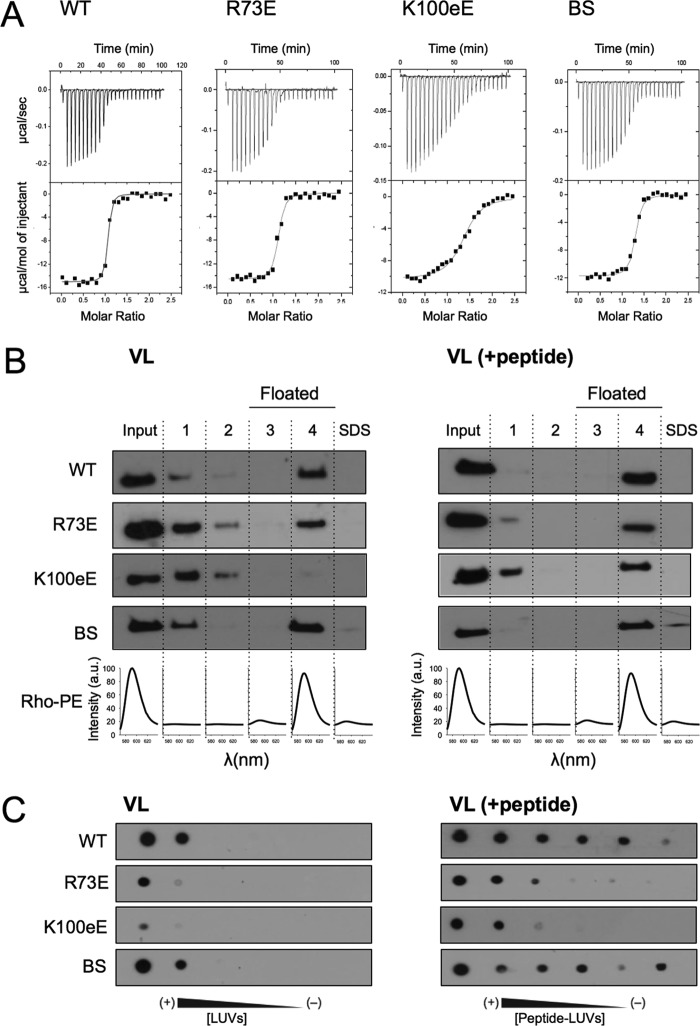
FIGURE 3.
Functional characterization of 4E10 mutants: binding to epitope peptide. A, binding isotherms of the MPER(664–690) epitope peptide to Fab 4E10 examined by ITC. The upper panel indicates the heat released upon consecutive injections of 10 μl of peptide solution (40 μm) into Fab (3 μm) in the calorimeter cell, and the lower panel indicates the integrated heats (symbols) and non-linear least squares fit (solid line) to the data using a one site binding model with the program ORIGIN 7.0. The thermodynamic parameters of binding are displayed in Table 1. Each titration was carried out once. B, flotation experiments in the presence of VL LUVs (left panel) or VL LUVs containing MPER(671–693) peptide (right panel). C, recognition of VL LUVs (left panel) or VL LUVs containing MPER(671–693) peptide (right panel) as determined by dot blot analysis. 2-fold serial dilutions of 500 μm LUVs and 10 μm peptide was spotted on the filters. BS, negative control with a double Ser to Ala substitution at positions 28 and 30 of the heavy chain.