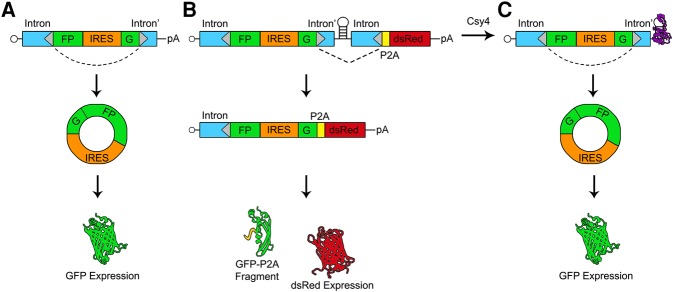
FIGURE 1.
Schematic of splicing patterns and reporter outputs for circGFP and circGFP-CD. The RNAs derived from the circGFP (A) and circGFP-CD (B,C) plasmids are expressed from the CMV promoter and are capped in their linear isoforms. Donor and acceptor splice sites are represented by gray triangles, and the dotted lines indicate the expected predominant splicing patterns. (A) The precursor circGFP transcript contains a split-GFP cassette flanked by intron sequences that have been engineered to contain inverted repeat sequences (blue boxes). The GFP fragments are separated by an EMCV IRES such that upon RNA circularization, full-length GFP is expressed. (B) The circGFP-CD precursor RNA contains the full circGFP sequence followed by a Csy4 targeted hairpin and a P2A-dsRed cassette. In the absence of Csy4 (B), forward splicing is favored and dsRed is translated. A nonfluorescent N-terminal GFP fragment by-product is released via the P2A sequence. In the presence of Csy4 (C), Csy4 can cleave the RNA at the base of the hairpin stem, releasing the forward splice acceptor and P2A-dsRed cassette. Csy4 remains associated with the cleaved hairpin, stabilizing the RNA and allowing back-splicing to occur. The IRES subsequently drives full-length GFP expression from the circular RNA.