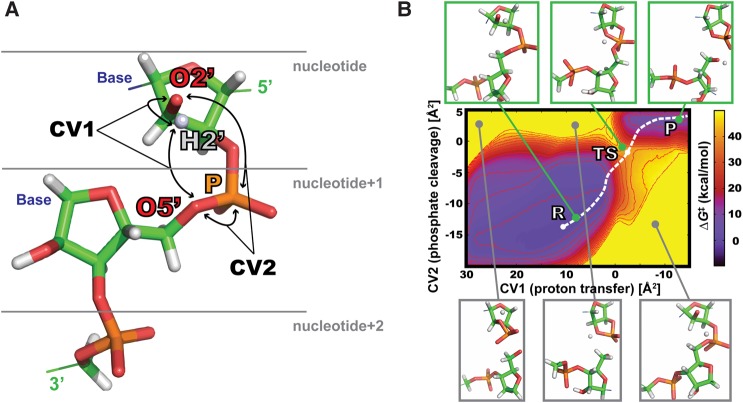
FIGURE 1.
(A) Selection of QM region and definition of collective variables (CVs). QM region for a particular nucleotide contained ribose ring (reactive 2′-OH group), phosphate with ribose ring of nucleotide+1, and phosphate moiety of nucleotide+2. Bases were described at MM level of theory (AMBER ff14). FES of phosphate cleavage reaction was defined by two CVs. CV1 described the proton transfer (difference of squared [O5′…H2′] and [O2′…H2′] distances), and CV2 described the phosphate cleavage (difference of squared [O5′…P] and [O2′…P] distances). (B) FES of phosphodiester backbone cleavage for G4 nucleotide from uGAAAg tetraloop. Snapshots in boxes show different conformation and reaction states of QM atoms defined by specific values of CVs. The proton from 2′-OH is represented as unbound to be clearly visible. Green boxes show geometries close to R, TS, and P states, whereas gray boxes display structures energetically penalized by restraints (see Supplemental Material for details). The white dashed line marks the estimated reaction coordinate.