FIGURE 3.
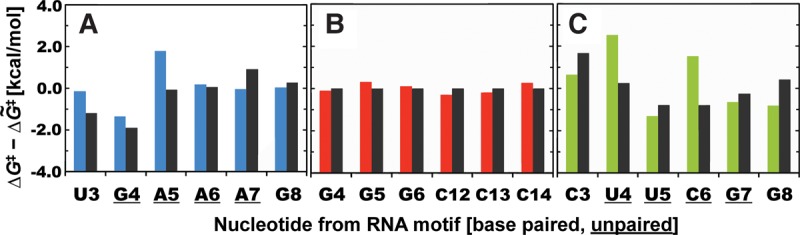
Comparison between computed and experimentally derived cleavage barriers for nucleotides from three different motifs. Correlation between computed ΔG‡ barriers (with specific color for each motif) and barriers derived from experiments (black, pseudo free energies, see Materials and Methods for details) for nucleotides from uGAAAg tetraloop (A), GC-duplex (B), and cUUCGg tetraloop (C). Both computed and experimentally derived ΔG‡ barriers are displayed as deviations from the corresponding median value. Note that in-line probing data for a uniform GC-duplex are not available, but equivalent G–C base pairs are expected to have similar ΔG‡ barriers (deviations from the median are set to zero in the figure). Unpaired nucleotides from tetraloops are underlined (see Supplemental Fig. S1 for nucleotide labeling).
