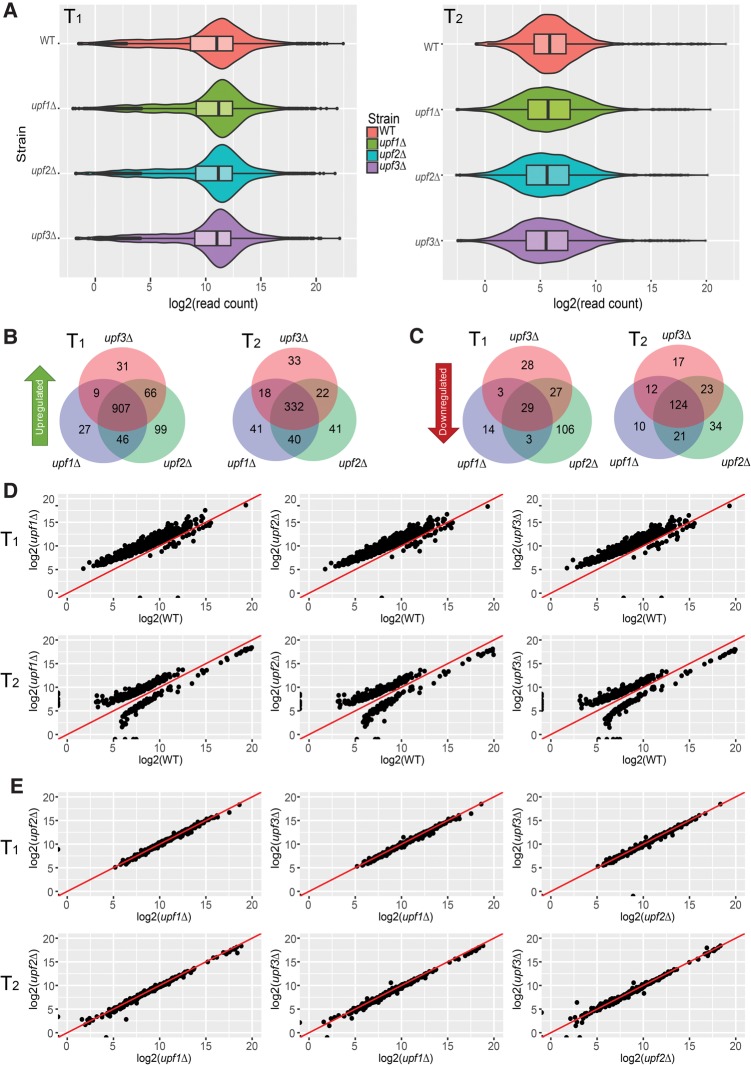
FIGURE 1.
Upf1, Upf2, and Upf3 regulate the same set of transcripts in yeast. (A) RNA-seq libraries from WT, upf1Δ, upf2Δ, and upf3Δ strains display comparable overall read count distributions for both transcriptome 1 (T1; left panel) and transcriptome 2 (T2; right panel). Violin and box-plots were used to visualize the average sequence reads distribution of the transcriptomes of the indicated strains from three independent experiments. (B,C) Transcripts up- and down-regulated in upf1Δ, upf2Δ, and upf3Δ strains show significant overlap. Transcripts up- or down-regulated in each UPF deletion strain were identified by comparisons to the WT strain. Venn diagrams were used to display the relationships among the sets of transcripts that are up-regulated (B) and down-regulated (C) in T1 or T2 of upf1Δ, upf2Δ, and upf3Δ strains. (D) All three UPF deletion strains display similar genome-wide expression patterns. Scatterplots were used to compare the read count values of differentially expressed transcripts between WT and upf1Δ, upf2Δ, or upf3Δ strains. The vast majority of differentially expressed transcripts in UPF deletion strains showed up-regulation and a small number of transcripts showed down-regulation. The y = x line is shown in red. (Top panel) Pairwise comparisons of the expression levels between WT and each UPF deletion strain for 936 differentially expressed transcripts from transcriptome 1. (Bottom panel) Pairwise comparisons of the expression levels between WT and each UPF deletion strain for 456 differentially expressed transcripts from transcriptome 2. (E) Transcripts commonly regulated by NMD each have virtually identical expression values in upf1Δ, upf2Δ, or upf3Δ strains. As in D, scatterplots were used to compare the read count values of NMD-regulated transcripts between upf1Δ and upf2Δ, upf1Δ and upf3Δ, and upf2Δ or upf3Δ strains. (Top panel) Differentially expressed transcripts from transcriptome 1. (Bottom panel) Differentially expressed transcripts from transcriptome 2.