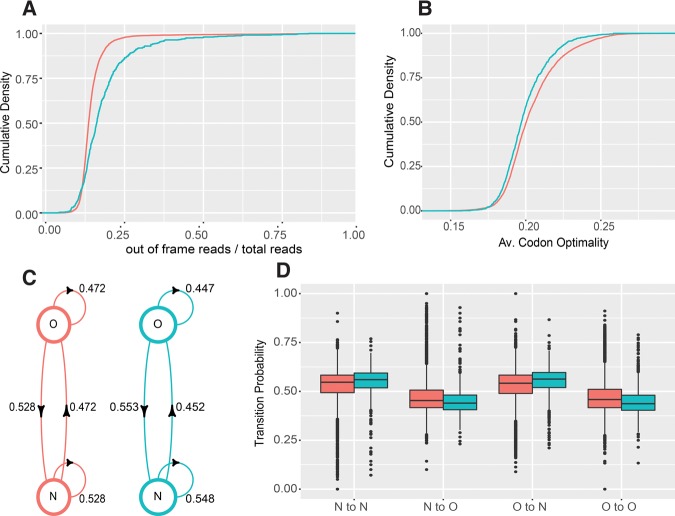
FIGURE 6.
NMD substrates have lower translation fidelity and lower codon optimality. (A) Cumulative density plots of in-frame read ratios over total reads derived from ribosome profiling data of WT cells for intron-lacking NMD substrates (n = 746, blue) and non-NMD substrates (n = 4633, red). (B) Cumulative density plots of mean codon optimality scores for two sets of transcripts shown in A. (C) Mean transition probabilities of a two-state discrete time Markov chain between optimal (O) and nonoptimal (N) codons for intron-lacking NMD substrates (blue) and non-NMD substrates (red). (D) Distributions of Markov chain codon transition probabilities for intron-lacking NMD substrates (blue) and non-NMD substrates (red). Plots in A were derived from the ribosome profiling data of Young et al. (2015), and plots in B, C, and D were based on codon optimality assignments and scores published by Pechmann and Frydman (2013). Two-sample KS test P-values are described in the Results section.