FIGURE 3.
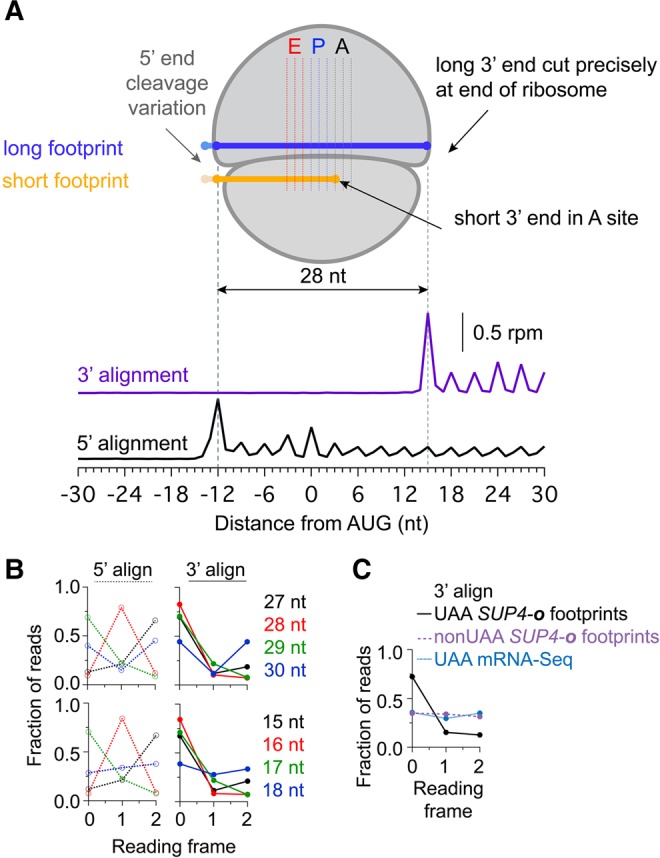
Analysis of mapped reading frame in 3′UTRs. (A) Schematic diagram of ribosome associated with long or short footprint reads. Short reads are always observed such that the 3′ end of the mRNA is positioned at the first nucleotide position of the A site. For reference, data underneath is average ribosome occupancy of long reads aligned by either 5′ or 3′ ends at start codons. Data are taken from the dom34Δ* SUP4-o strain on genes with non-UAA stop codons. This analysis also shows that the 3′ end of footprints maps more precisely to a single reading frame than the 5′ end (leading to reduced blurring of peaks at each codon). (B) Fraction of 5′ (left, dotted) or 3′ (right, solid) read ends mapping to each reading frame for long (top) and short (bottom) reads. Data derived from the dom34Δ* SUP4-o strain for 3′UTRs preceded by a UAA stop codon and no subsequent in-frame stop codons. (C) Fraction of 3′ read ends mapping to each reading frame for pooled short (15–18 nt) reads. Footprint reads from the dom34Δ* SUP4-o strain in cases where 3′UTRs are preceded by a UAA stop codon at the end of the ORF and include no subsequent in-frame stop codons are shown (black line) to preferentially occur in frame 0. In contrast, footprint reads from the same strain in cases with a non-UAA stop codon (purple line) are found equally well in all three reading frames. Similarly, mRNA-seq reads derived from the dom34Δ* strain for 3′UTRs preceded by a UAA stop codon at the end of the ORF and no subsequent in-frame stop codons in the 3′UTR (blue line) are equally distributed in all three frames.
