FIGURE 6.
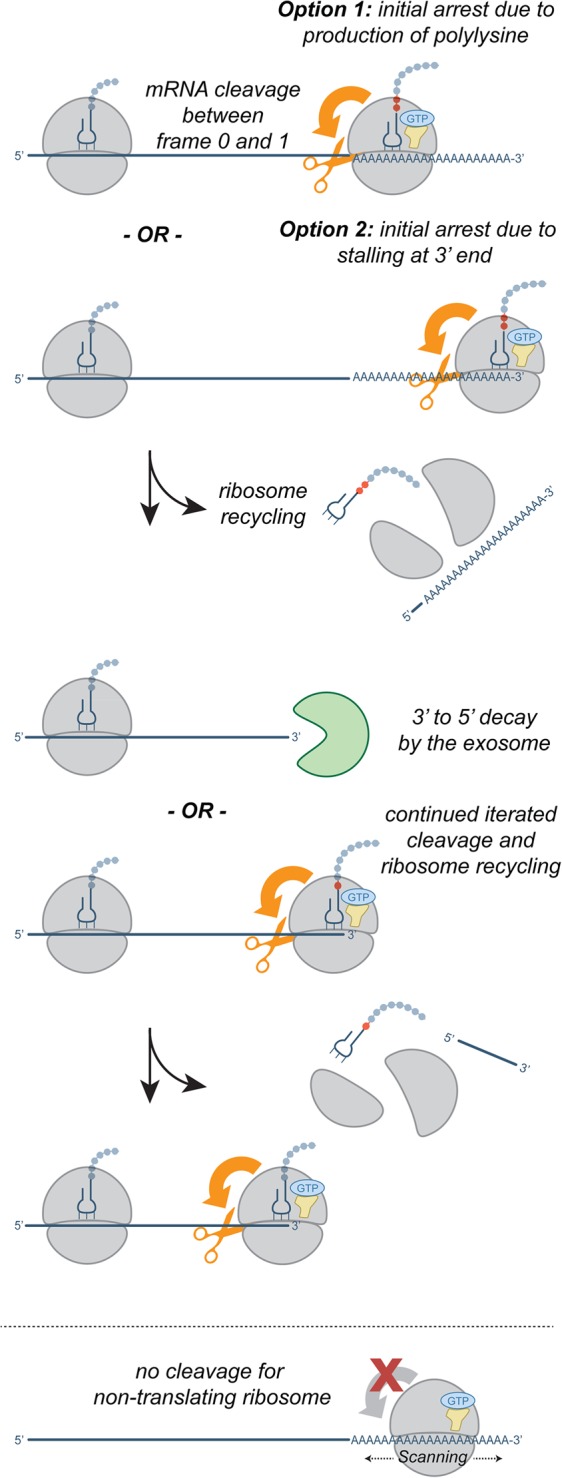
Model for translation-coupled mRNA cleavage during NSD/NGD. Ribosomes that translate the poly(A) tail and stall (top 2 rows) in the poly(A) tail (option 1) or at the end of the transcript (option 2). Stalled ribosomes recruit an endonuclease that cleaves the message upstream of the stalled ribosome (orange arrow). Upstream ribosomes in turn stall at these truncated ends, propagating the cleavage iteratively upstream. Once begun, this repetitive process may be interrupted by binding of the exosome to a truncated end. It can then carry out processive 3′–5′ decay until the transcript is degraded or the exosome collides with an upstream ribosome. Nontranslating ribosomes, such as those that fail to recycle properly at stop codons, do not bring about this iterative degradation pathway (bottom row).
