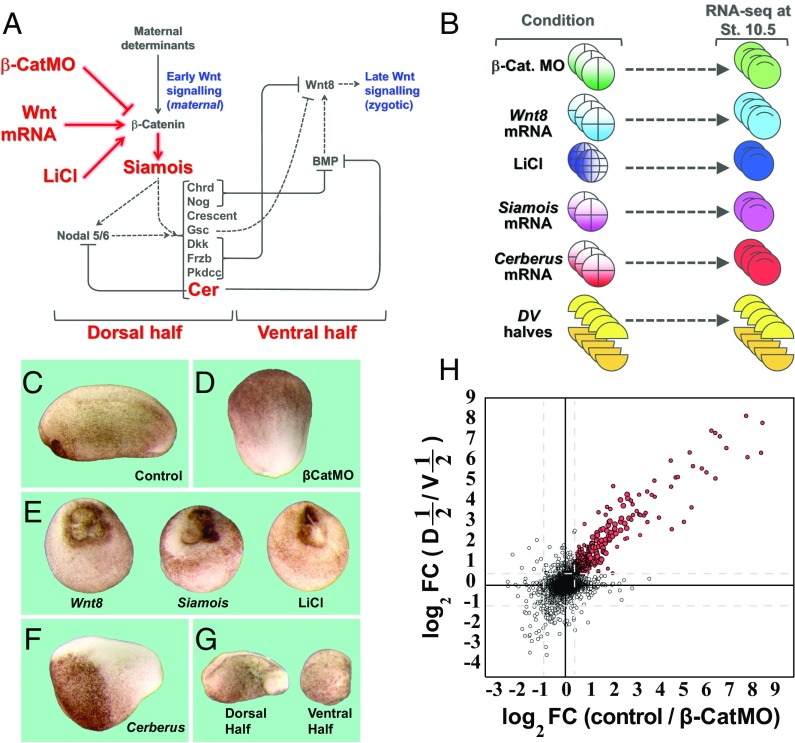
Fig. 1.
RNA-seq libraries for transcriptome analysis of early β-catenin targets. (A) Diagram showing the early β-catenin–dependent gene network on the dorsal side and the zygotic Wnt8 signal that arises later on the ventral side. The embryonic manipulations that were subjected to RNA-seq in the present study are indicated in red. (B) For RNA-seq, embryos were injected with β-catenin MO, Wnt8, Siamois, or Cerberus mRNA, treated with LiCl, or cut into dorsal and ventral halves at stage 8. All embryos were harvested at stage 10.5 for RNA extraction and RNA-seq. Some embryos were also analyzed at blastula stage 9. (C–G) Phenotypes of embryos subjected to manipulations in this study. Note that β-CatMO–injected embryos lacked all traces of a dorsal axis whereas Wnt8, Siamois mRNA microinjected, and LiCl-treated embryos were all radially dorsalized. Cerberus mRNA–microinjected embryos lacked endomesoderm and consisted mostly of anterior neural tissue and cement gland. Ventral half-embryos lacked dorsal axes whereas dorsal half-embryos developed a complete axis. (Magnification: 8×.) (H) Scatter plot comparing the log2 of fold change (FC) in RPKM upon β-catenin knockdown with that of dorsal halves/ventral halves. Shown here are 40,157 transcripts; red dots indicate transcripts decreased by β-catenin MO (by at least 1.4-fold in average of three experiments) and enriched in dorsal halves compared with ventral halves (by at least 1.5-fold in average of five experiments). Full datasets are provided in Dataset S1. Cer, Cerberus; D, dorsal half; St., stage; V, ventral half.