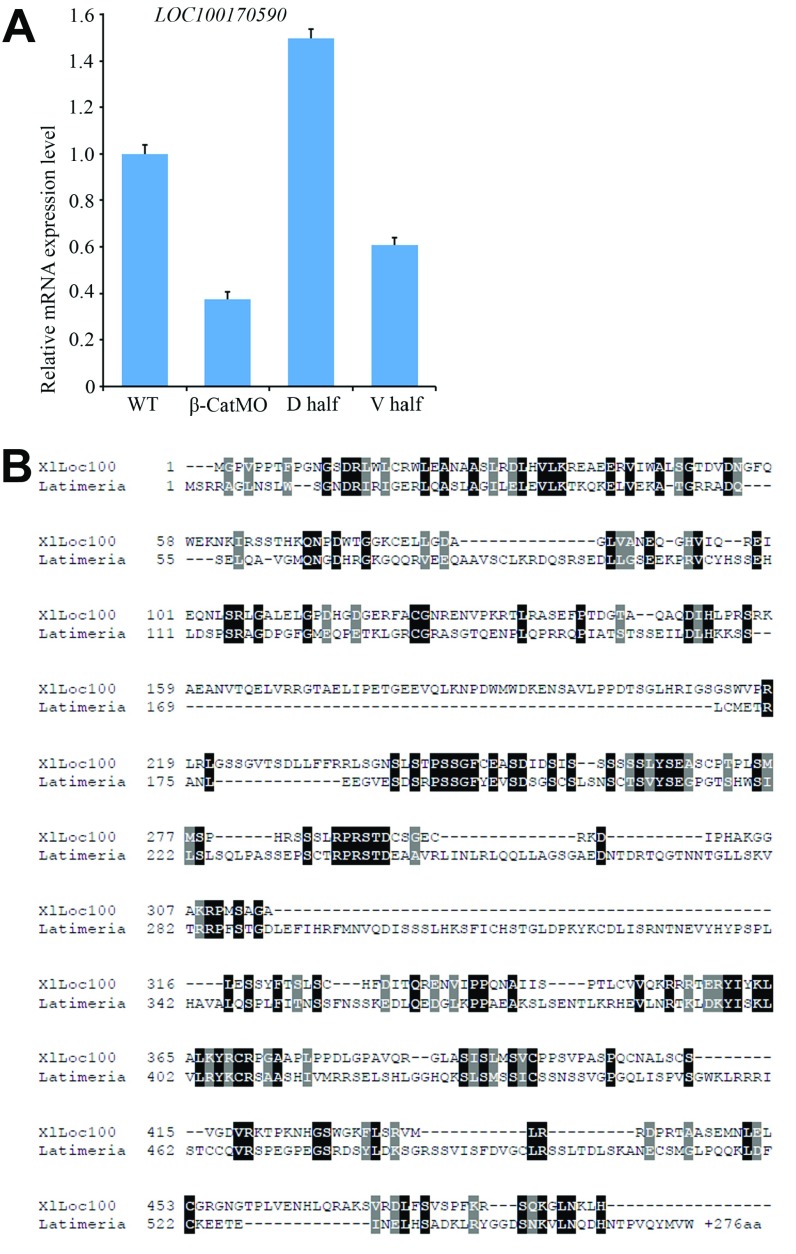
Fig. S2.
qRT-PCR validation and sequence homology of LOC100170590. (A) β-catenin–dependent regulation and D-V enrichment of LOC100170590 was validated by qRT-PCR analysis. β-CatMO decreased LOC100170590 mRNA levels compared with uninjected WT controls (WT), and expression in dorsal half-embryos (D half) was significantly higher than in ventral half-embryos (V half). (B) Amino acid sequence comparison between Xenopus laevis Loc100170590 and Latimeria chalumnae XP_005988953.1 (a coelacanth fish) revealed a 27% degree of identity. The coelacanth gene has been annotated as possible Dapper homolog-like 2, but XlLoc100170590 lacks critical Dapper-like features, such as a PDZ domain and a leucine zipper. The sequence alignment was generated by using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/) followed by BoxShade (embnet.vital-it.ch/software/BOX_form.html). Black and gray shaded boxes indicate identical and similar amino acid residues, respectively. There were no significant homologies with mammalian genomes.