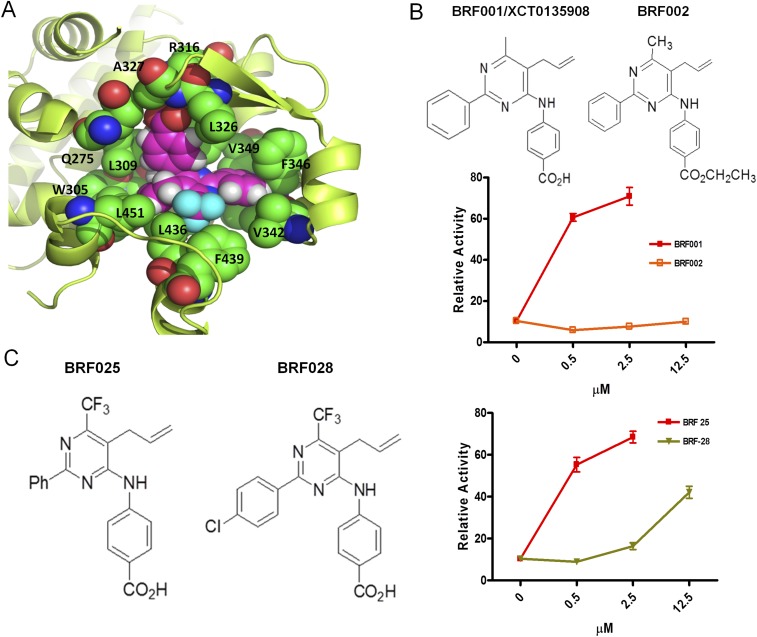
Fig. S5.
Structure of BRF110 docked in the X-ray structure of RXRα (PDB ID code 1MV9). (A) In silico docking simulations using the Glide “protein and ligand preparation” module. BRF110 is represented by magenta spheres, and neighboring amino acids are shown as green spheres. The surrounding protein is shown in yellow cartoon representation. The binding pocket of RXRα is purely hydrophobic except for the side chain of R316, which forms a salt bridge with the carboxylic acid of BRF110. The carboxylic acid also forms two hydrogen bonds with A327 and a neighboring water molecule. The hydrophobic enclosure of BRF110 is exemplified by its perfect shape complementarity with the L-shaped binding pocket of RXRα through favorable van der Waals interactions. Α271, I268, C269, and Α272 seal the hydrophobic pocket from the top and thus are hidden for clarity. (B) Modification of the carboxy group of various BRF compounds blocks activation of Nurr1:RXRalpha heterodimers. Examples of BRF001 (XCT0135908) and BRF002 are shown. (C) Disturbance of the electron cloud of the phenyl ring of various BRF compounds by the substitution of a hydrogen (BRF025) atom with a halogen (BRF028) atom decreased affinity for the RXRα-binding pocket.