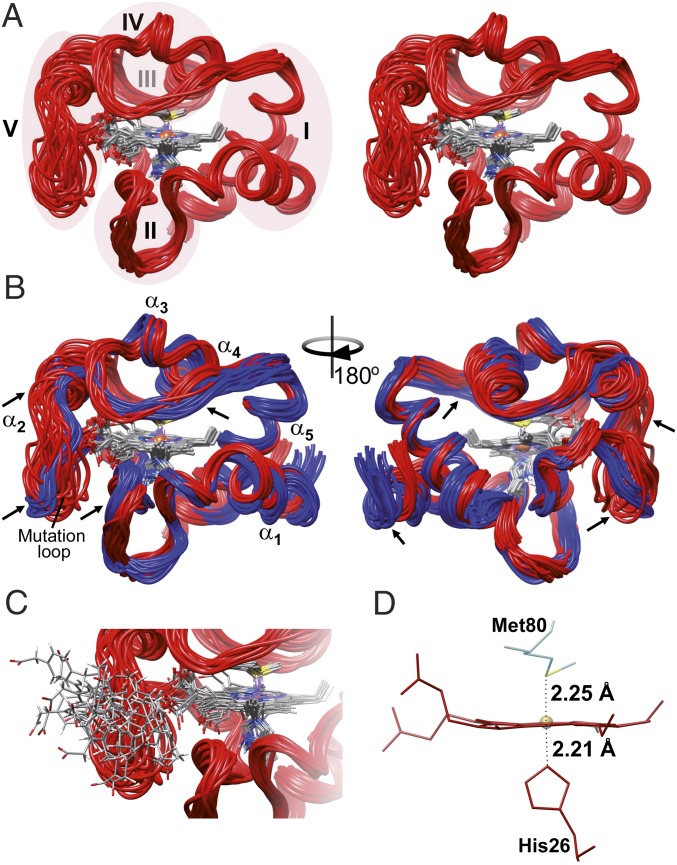
Fig. 2.
NMR solution structure of the Y48pCMF variant of Cc. (A) Stereoview ribbon representation of the 20 best conformers of Y48pCMF Cc. Heme group atoms are displayed for all conformers. Ribbons are colored in red, whereas atoms from the heme group are colored following the CPK (Robert Corey, Linus Pauling, and Walter Koltun) color scheme. Foldons of Y48pCMF Cc are shadowed and marked with roman numerals, except for foldon III, which is located behind foldon IV. (B) Comparison between the NMR solution structures of WT Cc (PDB ID code 1J3S) (33) and Y48pCMF Cc (this work). The ribbon for WT Cc is in blue. The five α-helices of both Cc species, as well as the mutation-containing loop of Y48pCMF Cc, are marked. Arrows point to the regions on the Y48pCMF Cc ribbon with substantial structural changes compared with the WT form. (C) Detailed view of the loop harboring the pCMF48 residue. pCMF48 atoms follow the CPK color scheme. Protein structures are presented by UCSF Chimera software (41). (D) Detail of the heme group and axial ligands. Labels display iron-to-axial ligand distances for the Y48pCMF mutant obtained from the EXAFS analysis (SI Appendix, Fig. S5).