Table S1.
Calculated abundances of peptides representing variously digested forms of the Ser66 and Ser458 loci of DET1
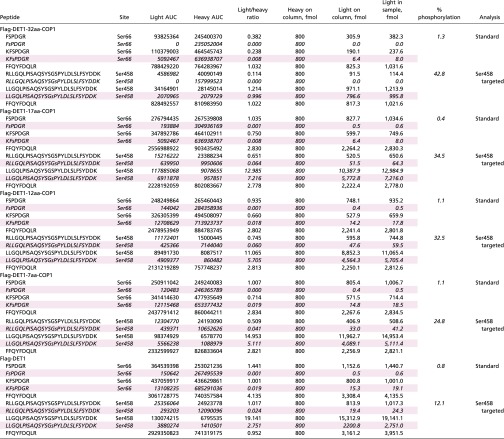 |
Peptides from the Ser66 locus were analyzed by a standard LC-MS/MS method, and those from the Ser458 site were analyzed by a targeted method specially tailored to favor hydrophobic, later-eluting features. An unmodified Det1 peptide (FFQYFDQLR) with characteristics amenable to both methods was quantified in each as a measure of consistency. Rows representing phosphorylated peptides are shaded pink. Values reported for the percent of phosphorylation were made in a locus-specific manner as Det1phos/Det1total. For the Ser66 locus, Det1total is the summed abundance of all four peptides quantified from this locus, including the two unmodified species (FSPDGR, KFSPDGR) and the two pSer66 phosphopeptides (FsPDGR, KFsPDGR; the lowercase “s” denotes the phosphorylated residue). Det1phos is the summed abundance of the two pSer66 phosphopeptides. At the Ser458 locus, Det1total is the summed abundance of the four peptides measured from this locus, i.e., the unmodified and phosphorylated versions RLLGQLPISAQSYSGSPYLDLSLFSYDDK, LLGQLPISAQSYSGSPYLDLSLFSYDDK and RLLGQLPISAQSYSGsPYLDLSLFSYDDK, LLGQLPISAQSYSGsPYLDLSLFSYDDK. Italicized sequences correspond to phosphorylated peptides.
