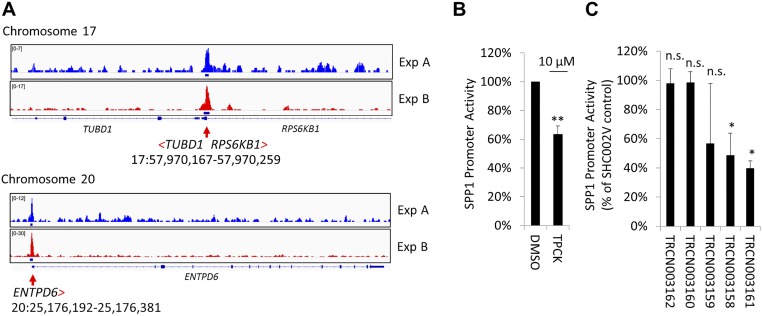
Fig. S6.
FAK ChIP-seq in WM858 melanoma cells. We sought to determine whether FAK associates with specific promoters in WM858 cells by using ChIP-seq. Three FAK antibodies detected 182 partially overlapping peaks in six ChIP-seq experiments, and 74 were detected in two independent experiments. (A) Exemplary promoter regions identified with high fidelity and within 2 kb of the transcription start site are shown and include the RPS6KB1 promoter. Based on 182 peaks detected, we determined the following consensus binding sequence: GCGC[AC]TGCGC. This binding sequence is highly similar to the optimal binding site for nuclear respiratory factor 1 (NRF1), (T/C)GCGCA(C/T)GCGC(A/G), which activates the expression of key metabolic genes. (B) To test whether the RPS6KB1 gene product, ribosomal S6 Kinase 1 (S6K1), helps drive SPP1 promoter activity, we exposed WM858 Dual-Glo cells to N-p-Tosyl-l-phenylalanine chloromethyl ketone (TPCK), a p70S6 kinase inhibitor. TPCK inhibited SPP1 promoter activity by ∼40%. SPP1 promoter activity after overnight TPCK treatment of WM858 cells is shown (mean ± SD; n = 3; **P < 0.001). (C) Likewise, two of five RPS6KB1-directed TRC shRNAs significantly inhibited SPP1 promoter activity. Effects of various TRC clone-derived RPS6KB1 shRNAs on SPP1 promoter activity are shown (mean ± SD; n = 2; *P < 0.05). n.s., not significant.