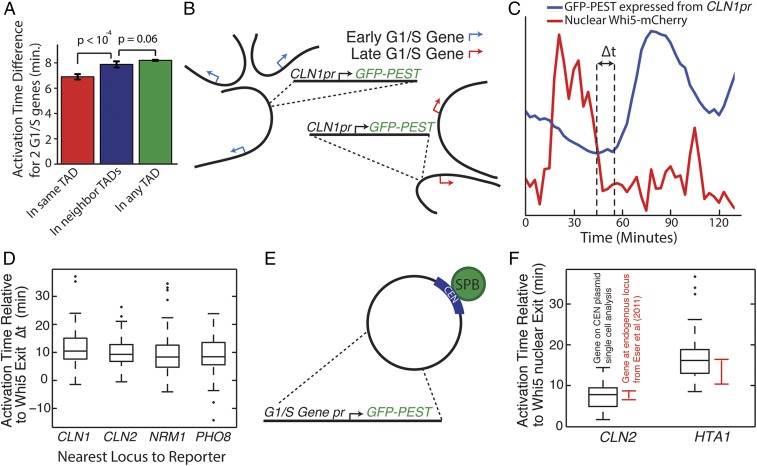
Fig. 3.
Cell cycle-dependent transcriptional activation is independent of gene nuclear location. (A) The timing of transcriptional activation in the cell cycle for all genes regulated by the SBF and MBF G1/S transcription factors was calculated in Eser et al. (35). Genes within the same TAD are activated at similar times compared with genes in neighboring domains. (B) Schematic of G1/S gene translocation experiment in which a construct containing an early gene promoter, CLN1pr, regulating a destabilized GFP, is integrated next to either an early activated gene or a late activated gene. (C) Transcriptional activation time in the cell cycle was calculated relative to the exit of the transcriptional inhibitor Whi5 from the nucleus, which marks the point of commitment to the cell cycle in late-G1 phase. Cells express a Whi5–mCherry fusion protein from the endogenous locus. (D) CLN1pr-GFPpest was integrated at the CLN1, CLN2, NRM1, and PHO8 loci, but this did not change the distribution of promoter activation times (P > 0.05 all comparisons). (E) Schematic for experiment comparing gene activation times for promoter GFP constructs expressed from CEN (centromere) plasmids, which localize near the spindle pole body (SPB). (F) The relative timing of transcriptional activation is similar for an early activated (CLN2) and a late activated (HTA1) gene expressed from either CEN plasmids or the genome. Data for transcriptional activation from the genomic loci are from Eser et al. (35).