Figure 2. NTP exit pathways via secondary pore from molecular dynamics simulations.
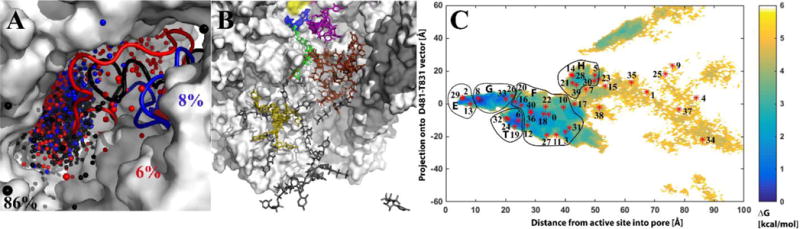
Secondary pore exit pathways based on NTP center of mass positions (shown as black, red, and blue spheres) from 100 RAMD simulations projected onto the initial structure of Pol II. The trigger loop is shown in cartoon representation and colored corresponding to each pathway. Percentages indicate the relative occurrence of each pathway (A). Macrostates from MSM shown as UTP orientations within the secondary pore of the initial structure of Pol II. Conformations grouped into superstates are colored as follows: E, blue; G, green; T, purple; F, brown; H, tan; and bulk, grey (B). PMF for NTP positions in the secondary pore from snapshots of unbiased simulations as a function of the distance from the active site and projection onto the Cα D481→Cα T831 vector. Binding states used in the kinetic model (Fig. 3) are indicated as letters (C).
