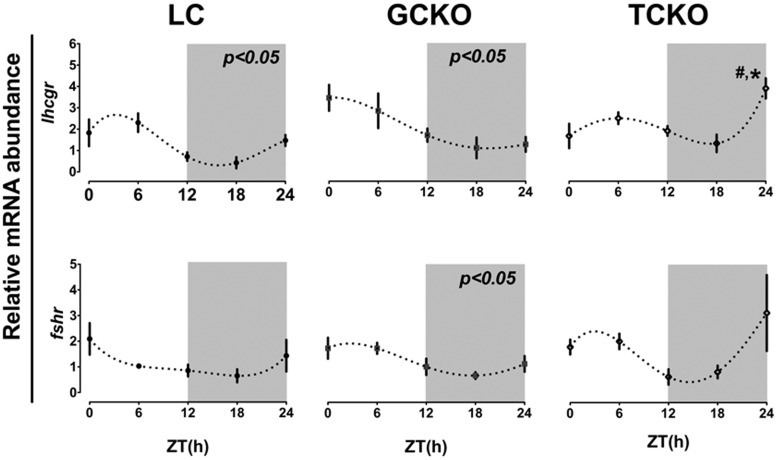
Figure 6. Conditional KO of Bmal1 in TCs alters rhythms of LHR mRNA abundance in the ovary.
Rhythms of Lhcgr and Fshr expression in whole ovary tissue recovered from LC (n = 3), GCKO (n = 3), and TCKO (n = 3) transgenic mice. In both LC and GCKO mice we detected a significant diurnal rhythm of Lhcgr mRNA abundance (P < .05 for both) that was abolished in TCKO transgenic mice. There was a marked increase in Lhcgr expression at ZT24 in TCKO mice (#, P < .05) when compared with the same time point in both LC and GCKO mice. A low amplitude oscillation of Fshr mRNA expression was detected in GCKO transgenic mice with a peak in the morning (ZT1.99). Daily variation of Fshr mRNA abundance was not significant in either LC or TCKO transgenic mice. Data are presented as mean ± SEM. n = 3 ovaries from 3 different mice per time point. #, P < .05 GCKO vs TCKO; *, P < .05 LC vs GCKO or TCKO. Only those rhythms labeled directly with a P value were considered significant by CircWave analyses. Values for target mRNA abundance were calculated using the ΔΔCT method as described in Materials and Methods, with β-actin as the reference gene. Data were fit to a nonlinear regression (see Materials and Methods).