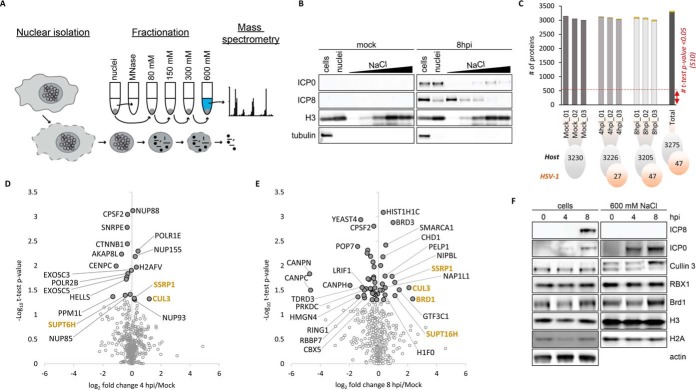
Fig. 4.
Analysis of proteins accumulated in chromatin fraction during HSV-1 infection. A, Workflow of high salt fractionation of chromatin-associated proteins. Purified nuclei were digested with MNase enzyme to separate chromatin into soluble and insoluble fractions. Subsequently, the insoluble fractions were washed with increasing concentrations of salt (80–600 mm NaCl). The high salt fraction, which solubilized the majority of nucleosomes was subjected to MS-analysis. B, Western blots of selected viral (ICP0, ICP8), and host (H3, tubulin) proteins showing their abundance at mock and 8 hpi in the whole HeLa cell extract, whole nuclei extract, MNase treated fraction, and individual fractions over the salt gradient (80, 150, 300, and 600 mm NaCl). C, Number of quantified proteins for host (gray) and virus (orange) at each time point and biological replicate analyzed in the high salt fraction (600 mm NaCl). D, Volcano plot representing the fold change and significance of host protein abundance at 4 hpi versus mock and (E) 8 hpi versus mock. The standard significant threshold (t test p value < 0.05) transformed in -Log10 is 1.30. 'Light gray dots' highlight t test p value > 0.05. Only proteins involved in chromatin organization processes were highlighted (annotation retrieved from MetaCore database; False Discovery Rate (FDR < 0.05). F, Western blots of selected host proteins representing their abundance at 0, 4 and 8 hpi in the whole cell extract and the high salt fraction (600 mm NaCl). BRD1 and Cullin 3 accumulated in the high salt fraction at 8 hpi.