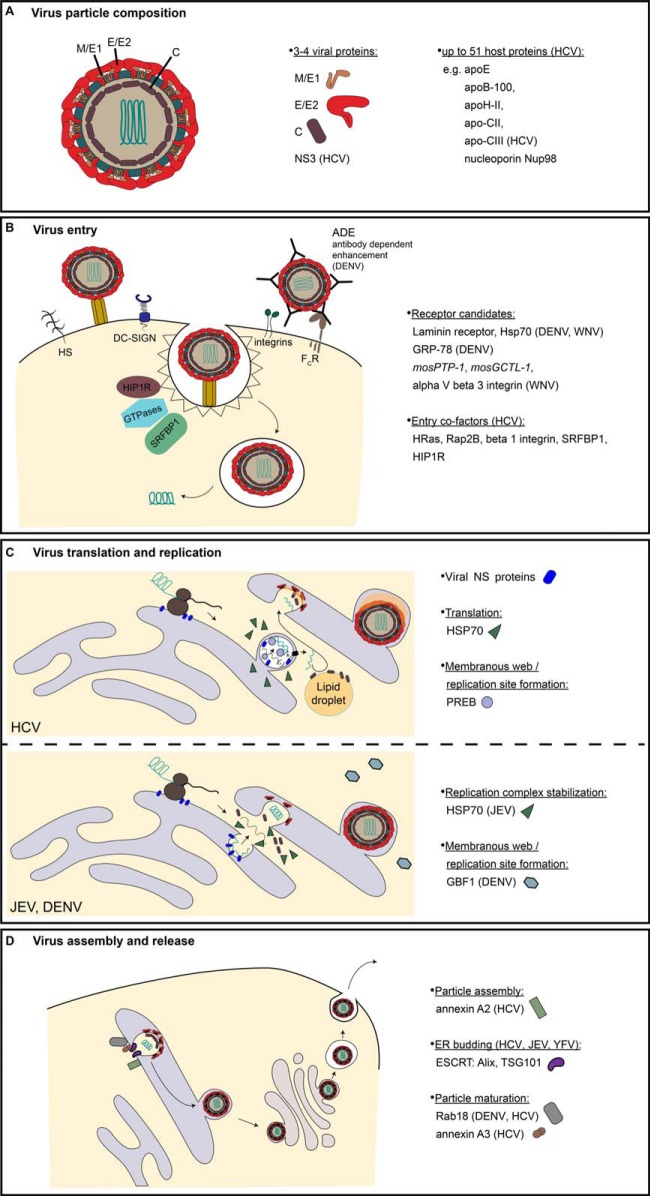
Fig. 3.
Protein interactions during the flavivirus life cycle elucidated by MS-based proteomics. (A) Flavivirus particle composition. Flaviviridae particles are composed of a capsid/core (C) surrounded positive strand RNA genome and enveloped by a host derived lipid bilayer in which the viral transmembrane domain glycoproteins M/E1 and E/E2 are embedded. Additionally, the virions may comprise of nonstructural proteins and host proteins. (B) Virus host protein interactions during entry. Flaviviridae members penetrate the host cell by receptor-mediated endocytosis and fusion in endosomal compartments. MS-based proteomics discovered the listed interaction partners of viral glycoproteins as receptor candidates. SILAC-approaches further revealed secondary interactors of HCV through co-IP MS of the HCV receptor CD81 (listed as entry cofactors). (C) Virus host protein interactions during translation and replication. The Flaviviridae viral genomes translate and replicate in the cytosol in close association with ER membranes, which are remodeled during infection. MS-based proteomics identified heat shock protein 70 (HSP70) as host translation factor and prolactin regulatory element binding (PREB), HSP70, and GBF1 as host replication factors for the indicated viruses. Viral NS proteins forming the replication complex are depicted by a single blue symbol. (D) Virus host protein interactions during particle assembly and release. Particles are assembled at ER-derived membranes and exit the cell through the secretory pathway. MS-based proteomics revealed annexins, ESCRT components, and the GTPase Rab18 as host factors promoting particle assembly, ER budding, and maturation of the indicated Flaviviridae members. All panels show the assumed but not necessary experimentally confirmed localization of host factors identified by MS-based proteomics.