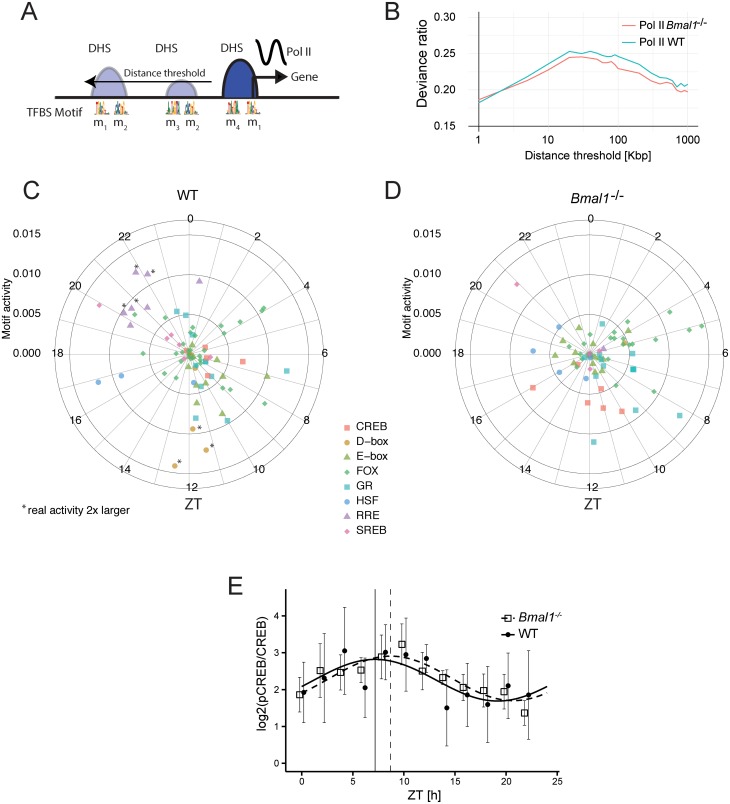
Fig 4. Distal DNase I Hypersensitive Sites (DHSs) help identify diurnally active transcription regulators.
A. Scheme of the linear model to infer active transcription regulators: transcription factor (TF) motifs in DHSs within a symmetric window around active transcription start sites (TSSs) are used to explain diurnal rhythms in transcription. B. Fraction of explained temporal variance (deviance ratio) in RNA polymerase II (Pol II) loading (at the TSS of all actives genes) for WT and Bmal1-/- mice, in function of the window size (radius) for DHS inclusion, shows a maximum at around 50 kb. Here, α = 0 was used in the glmnet (Materials and methods). C–D. Inferred TF motif activities for WT and in Bmal1-/- mice shown with amplitudes (distance from center) and peak times (clockwise, ZT0 at the top) using a window size of 50 kb. All 819 (WT) and 629 (Bmal1-/-) motifs (overlap is 427) with nonzero activities are shown. Note though that most activities are very small and cluster in the center. Certain families of TFs are indicated in colors (full results are provided in S4 Table). Radial scale for activities is arbitrary but comparable in C and D. E. Quantification of western blots for pCREB (Ser 133 phosphorylation) and CREB in WT and Bmal1-/- genotypes (log2 (pCREB/CREB)). Nuclear extracts from four independent livers were harvested every 2 h. Both genotypes showed a significant oscillation (p < 0.05, harmonic regression) of the mean signal from the four mice. Though the peak time in Bmal1-/- mice is delayed by 1.8 h, the comparison of the rhythm in the two genotypes was not significant (p = 0.49, Chow test). Individual blots are shown in S7 Fig.