Abstract
The Protein Ontology (PRO) is the reference ontology for proteins in the Open Biomedical Ontologies (OBO) foundry and consists of three sub-ontologies representing protein classes of homologous genes, proteoforms (e.g., splice isoforms, sequence variants, and post-translationally modified forms), and protein complexes. PRO defines classes of proteins and protein complexes, both species-specific and species non-specific, and indicates their relationships in a hierarchical framework, supporting accurate protein annotation at the appropriate level of granularity, analyses of protein conservation across species, and semantic reasoning. In this first section of this chapter, we describe the PRO framework including categories of PRO terms and the relationship of PRO to other ontologies and protein resources. Next, we provide a tutorial about the PRO website (proconsortium.org) where users can browse and search the PRO hierarchy, view reports on individual PRO terms, and visualize relationships among PRO terms in a hierarchical table view, a multiple sequence alignment view, and a Cytoscape network view. Finally, we describe several examples illustrating the unique and rich information available in PRO.
Keywords: Protein ontology, proteoforms, protein complexes, post-translational modification, orthologs
1. Introduction
Aberrations in protein activities are a fundamental cause of human diseases. Pathological changes in the proteome may result from single amino-acid variations resulting from non-synonymous single nucleotide polymorphisms (nsSNPs) [1,2], abnormal isoforms arising from aberrantly alternative spliced mRNAs [3,4], changes in post-translational modifications (PTMs) [5,6], or changes in cooperative behavior of multiple proteins in a protein complex [7,8], as well as interdependencies of these mechanisms. With the advent of high-throughput proteomics technologies, our understanding of the protein composition of human cells in health and disease is expanding rapidly, especially when proteomics data are overlaid and analyzed along with genomic, transcriptomic and interactomic data in their biological context.
The Protein Ontology (PRO, proconsortium.org) [9] is a reference ontology for proteins and protein complexes in the OBO (Open Biological and Biomedical Ontologies) Foundry [10] that offers a research infrastructure for modeling biological systems and integrating existing and emerging experimental data.
PRO defines classes of proteins and protein complexes and indicates how these classes interrelate. For knowledge representation, PRO defines precise protein entities to support accurate annotation at the appropriate granularity and provides the ontological framework to connect all protein types necessary to model biology, in particular linking specific protein forms to particular complexes and particular functions in their biological context. For semantic data integration, PRO provides the ontological structure to connect—via specified relations—the vast amounts of protein knowledge contained in databases to support new hypothesis generation and testing.
Classes defined in PRO can be either organism-non-specific or organism-specific and range in granularity from protein family to proteoform classes (which account for the precise molecular form of a protein, including specification of sequence or splice variant and any post-translational modification or PTM [11]). Thus, it allows precise definition of protein objects and the specification of their relationships with each other.
1.1. PRO Framework
To model the various types of protein entities, we have formulated three sub-ontologies of PRO to represent: (i) protein classes of homologous genes, (ii) protein forms (proteoforms [11]) arising from single genes, including splice isoforms, mutation variants, and PTM forms, and (iii) protein complexes [9,12,13]. Protein terms in PRO are defined at multiple levels of granularity from the family level down to the isoform and/or modification level, allowing annotation at the most appropriate level given current knowledge. For example, as 14-3-3 proteins are encoded by several genes whose protein products may not be distinguishable in assays, they are represented by PR:000003237 for protein products of the 14-3-3 gene family. Similarly, when the protein is known to be the product of a given gene but the precise isoform is not known, then a gene-level PRO term covering all protein products is used (e.g., TP73, PR:O15350).
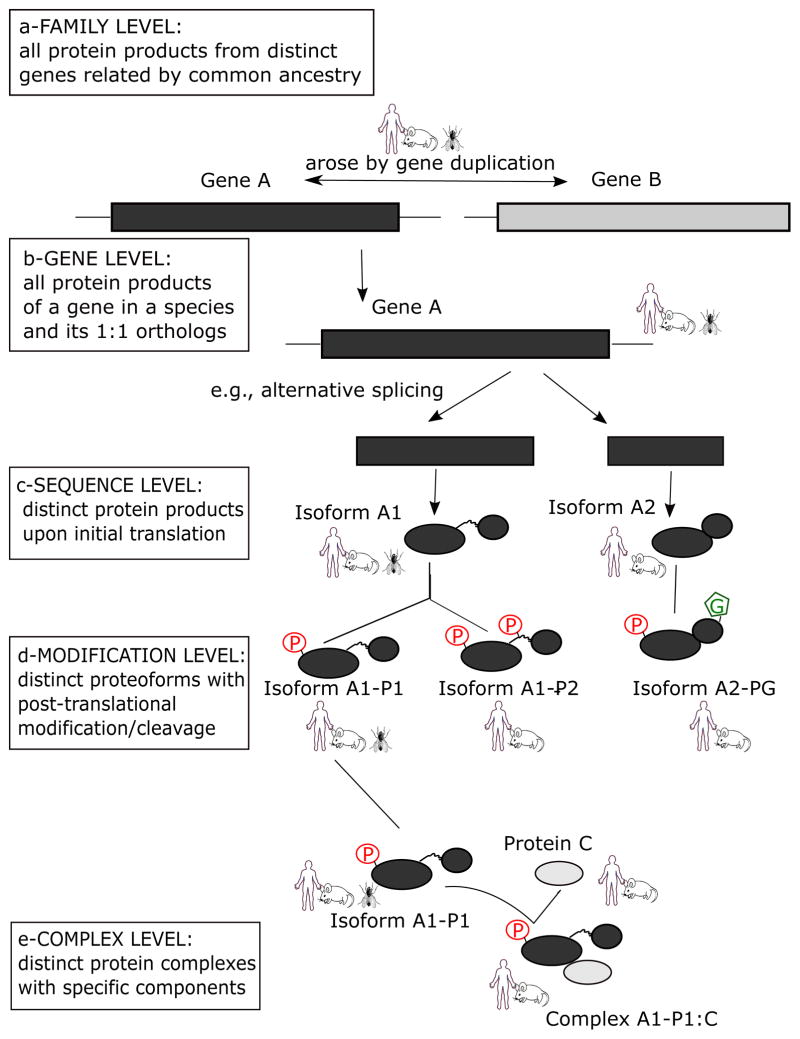
Figure 1 shows a schematic representation of the ontology, which is organized in different levels as follows:
Figure 1.
PRO Hierarchy. The diagram shows the different classes that can be represented in the ontology. From top to bottom: a-Family class includes all the protein products of evolutionary related genes at the homeomorphic level (e.g., all the protein products of Gene A and Gene B are in the same family class). These are conserved in a set of taxa (e.g. human, mouse, fly). b-Gene level includes all the protein products of a distinct gene in a species and its 1:1 orthologs. In PRO, human is the reference organism for vertebrates and all gene products of Gene A in human and its orthologs (e.g. mouse and fly) are in the same gene level class. Note that the gene is shown as a box because the gene structure (e.g. number and positions of introns) may differ between species. Species-specific organism-gene classes are children of the corresponding Gene level (e.g. mouse Gene A, and human Gene A are both members of the same Gene A class). c-Sequence level includes all the isoforms produced by initial translation. This example shows two protein classes isoforms A1 and A2, created by alternative splicing, where isoform A1 is conserved in human, mouse and fly species, and isoform A2 is observed only in mammals. Again, the species specific organism-sequence terms can be created. d-Modification level includes all post-translational modifications. Shown here are proteoforms of isoform A1: P1 (phosphorylated at a single site) and P2 (phosphorylated at two sites) and proteoforms of Isoform A2: P1 (phosphorylated at a single site) and PG (phosphorylated and glycosylated). e- Protein complex level defines complexes based on component subunits (with stoichiometry if known). In this case, proteoform isoform A1 P1 and protein C are components of complex A1P1:C.
Family: refers to the class of proteins translated from a specific set of ancestrally related genes. Proteins in this class can be traced back to a common ancestor showing homology over the entire length of the protein. The leaf-most nodes at this level are usually families comprising paralogous sets of gene products (of a single or multiple organisms). Figure 1 shows that gene A and gene B arose by gene duplication (paralogs) and that all protein products of gene A and gene B would be under the same family class in PRO. For example, in PRO the potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel protein class (PR:000000676) is defined as “A protein with amino- and carboxyl-terminal intracellular domains separated by a domain (common with other ion channels) containing six transmembrane helices (S1–S6) in which the last two helices (S5 and S6) flank a loop, called the pore loop, which determines ion selectivity. The N terminus has a conserved domain that is also present in other voltage-gated potassium and sodium channels. The carboxyl-terminal region contains the cyclic nucleotide-binding domain (CNBD). In addition, there is a structural element called the C-linker, the region connecting the CNBD to the S6 segment, which couples conformational changes in the ligand-binding domain to channel activation…. [PMID:16382102]”. This class includes the protein products of genes HCN1, HCN2, HCN3 and HCN4.
Gene: a PRO term at this level refers to a class of proteins translated from a gene related by 1:1 orthology in distinct organisms. Considering human as a reference, all protein products of Gene A in human and its 1:1 orthologs in Figure 1 would fall under the gene level class. The Gene A protein products from mouse and fly would also be included. Continuing with a real example, the HCN4 gene product (PR:000000708) is defined as “A potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel protein that is a translation product of the human HCN4 gene or a 1:1 ortholog thereof. [PRO:CNA].” This class currently includes the protein products of the HCN4 gene in rat, mouse, rabbit and human. The species-specific genes (e.g. the human version of Gene A in Figure 1, or mouse version of HCN4 in the example above) are children of the corresponding gene level terms. PRO uses the OMA orthology database [14] to map the organism gene to the corresponding gene level for selected model organisms.
Sequence: A PRO term at this level refers to the protein products with a distinct sequence upon initial translation. The sequence differences can arise from different alleles of a given gene, from splice variants of a given RNA, from alternative initiation or termination of transcription, and from ribosomal frame shifting during translation. One can think of this as a mature mRNA-level distinction. In Figure 1, isoform A1 (observed in human, mouse and fly) and isoform A2 (observed only in human and mouse) would be two different classes in PRO. Again, under each term the species-specific terms can be created, and these are called ortho-isoforms.
Modification: a PRO term at this level refers to the protein products derived from a single mRNA species that differ because of some change (or lack thereof) that occurs after the initiation of translation (co- and post-translational). This includes sequence differences due to cleavage and/or chemical changes to one or more amino acid residues. The example in Figure 1 shows two proteoforms of isoform A1 that differ in their phosphorylation state (single vs. doubly phosphorylated form). We have recently collected information about proteoforms of beta-catenin [15], specifically in relation to cancer. The phosphorylation state of beta-catenin influences its stability and its interacting partners. Phosphorylation of a specific set of residues is needed for its degradation. Phosphorylation of Ser-45 on human beta-catenin by casein kinase I (CKI) followed by the sequential phosphorylation of Thr-41, Ser-37, and Ser-33 phosphorylation by the glycogen synthase kinase, GSK3B, (PR:000035772) creates a recognition site for the ubiquitin ligase BTRC, which ubiquitinates beta-catenin, targeting it for degradation by the proteasome. Another modified proteoform of beta-catenin, phosphorylated on Tyr-654 (PR:000044478), has enhanced transcription-related functions. Fifteen proteoforms with distinct phosphorylation site combinations are described in PRO for human beta-catenin.
Complex: class of complexes with a specific defined subunit composition. PRO makes no distinction between complexes whose components are modified before or after complex formation. All complexes are grouped into the ‘complex’ category regardless of the specific components.
1.2. PRO and Interoperability with Other Protein-Related Resources
PRO collaborates closely with other ontologies and resources to maximize semantic interoperability. The organism-specific protein complexes defined in PRO extend the generic complexes described in the Gene Ontology (GO) Cellular Component Ontology (GO, [16]). The organism-non-specific complexes of GO provide parent terms for PRO’s organism-specific complex terms and provide the basis for connecting and comparing complexes between organisms. PSI-MOD [17] and the Sequence Ontology (SO) [18] are used to define protein classes of the modification category. PRO not only interoperates with ontologies, but with other resources as well. The ‘organism-gene’ level in PRO is equivalent to the UniProtKB entries for the specific protein sequences, including those of splice isoforms, arising from the gene represented [19]. Thus, PRO both incorporates UniProtKB and complements it by providing formal definitions for protein entities and placing the terms in an ontological context. Similarly, the Intact complex portal [20] contains protein complexes observed in specific major model organisms and these will be integrated into the ontological framework of PRO (ongoing effort).
1.3. PRO Applications
PRO has been employed in multiple studies including assisting in ontology building (especially in application ontologies), semantic integration, and functional annotation and ontological representation for proteoforms and complexes for proteomics studies. A few examples are listed below:
Used in combination with literature mining and curated databases to create a knowledge “map” for analysis of beta-catenin function in cancer [15].
Integrated in IDOBRU framework for ontological modeling of host-pathogen interactions using Brucella as the pathogen model [21].
Supporting GO annotation of specific proteoforms in model organism databases (e.g., in MGI [22] and PomBase [23]).
Supporting annotation of species-specific protein complexes in the Toll-like Receptor signaling pathway, relating both to their components and to species-independent families of complexes [12].
Used within the Neurological Disease Ontology, an ontology that represents aspects of neurological diseases that are relevant to their treatment and study [24].
Supporting concept recognition in CRAFT corpus [25].
Providing ontological framework for proteoforms in iPTMnet (http://proteininformationresource.org/iPTMnet/, see Chapter 16).
1.4. Scope of this chapter
In this tutorial, you will learn how to use the PRO resources, including i) searching/browsing the ontology and annotation; ii) analyzing proteoforms of a gene within and across species; iii) analyzing complexes; iv) saving/downloading data; and v) visualization of the ontology. Examples in this book chapter are from Release 49.0.
2. Materials
The PRO website is accessible at proconsortium.org.
Download: The ontology (pro.obo) and the annotation (PAF.txt) can be downloaded from the ftp site accessible from the PRO home page. The ontology is also available in OBO and OWL formats through the OBO Foundry (7) and Bioportal (8). For general documentation please see links on the PRO home page.
The pro.obo contains a stanza of information about each term. Each stanza in the obo file is preceded by [Term] and is composed of an ID, a name, synonyms (optional), a definition, comment (this section indicates the hierarchy level and may include evidence codes and other comments), cross-reference (optional) and one or more relationships to other terms (Figure 2).
The annotations to PRO terms are distributed in the PAF.txt file. To facilitate interoperability to the best extent this tab delimited file follows the structure of the gene ontology association (GAF) file. Please read the README file and the PAF guidelines.pdf in the ftp site to learn about the structure of this file. PRO terms are annotated with relations to other ontologies or databases. Currently used: Gene ontology (GO) [16] to describe processes, function and localization; Sequence ontology (SO) [26] to describe protein features; PSI-MOD (http://www.psidev.info/MOD) to describe protein modifications; Disease Ontology [27] to describe disease states; and Pfam [28] to describe domain composition.
-
Link to PRO. PRO identifiers are URIs, globally unique identifiers. The URIs have one of two forms:
http://purl.obolibrary.org/obo/PR_ddddddddd where d is a digit.
http://purl.obolibrary.org/obo/PR_ <uniprot accession>
For example, see http://purl.obolibrary.org/obo/PR_Q8CGC7.
Cytoscape. PRO features a Cytoscape [29] view that provides an interactive and visual interpretation of PRO terms and their relations. The view can be accessed via the Cytoscape icon displayed on PRO search results pages and entry reports. Cytoscape supports searches and customization of the layout and display; displays links to PRO entry pages, OBO stanzas and annotations; and allows saving of the network image in PNG format.
Figure 2.
Example of a PRO OBO stanza.
3. Methods
3.1. PRO Homepage
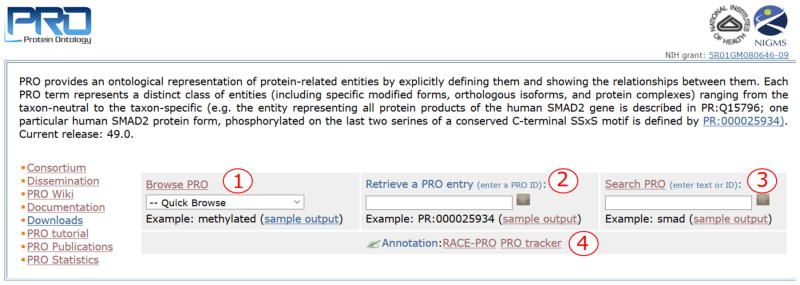
The PRO homepage (http://www.proconsortium.org/) (Figure 3) is the starting point of navigation through the protein ontology resources. The menu on the left side links to several documents and information pages, as well as to the ftp download page. The functionalities in the homepage include: (3.1.1) PRO browser, (3.1.2) PRO entry retrieval, (3.1.3) text search, and (3.1.4) annotation.
Figure 3.
PRO homepage. Main Functionalities include: 1) Browsing, 2) Retrieval, 3) Search, and 4) Annotation tools.
3.1.1. PRO Browser
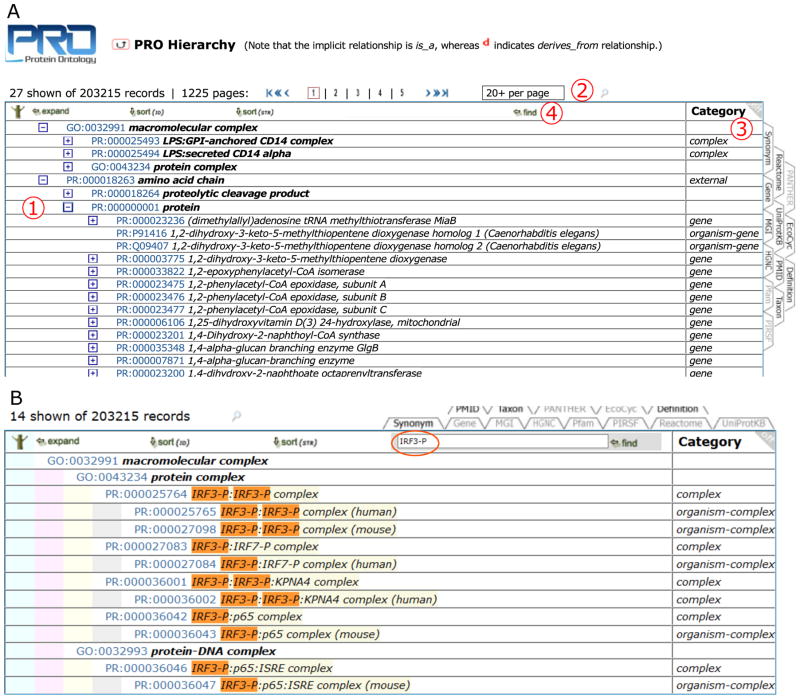
The browser is used to explore the hierarchical structure of the ontology (Figure 4). The icons with a plus and minus signs (Figure 4A, 1) are for expanding and collapsing nodes, respectively. Next to these icons is a PRO ID, which links to the corresponding entry report, followed by the term name. Unless otherwise stated, the implicit relation between nodes is is_a. The number of terms to be displayed in the hierarchy page can be managed from the result page number box (Figure 4A, 2). The column on the right (Figure 4A, 3) shows the level within the hierarchy for each term. This column is customizable, and additional information can be added by selecting other information tabs. Finally, to retrieve all terms matching a particular keyword, enter the word or phrase in the Find box (Figure 4A, 4) and all terms matching will be displayed in the hierarchy as shown in Figure 4B for terms containing IRF3-P, the phosphorylated form of interferon regulatory factor 3.
Figure 4.
PRO Browser. Navigate the ontology through its hierarchical view. A. In the PRO browser you can: 1) use plus and minus signs to expand/collapse terms, respectively; 2) change the number of terms displayed per page; 3) customize the information tabs; and 4) find terms matching a keyword or phrase. B. PRO browser view with terms containing IRF3-P, using the “find” functionality.
3.1.2. PRO Entry Pages
The PRO entry provides a report containing the ontological information and annotation available for a given PRO term. If you know the PRO ID you can use the “retrieve PRO entry” box in the homepage (Figure 3, 2) to get direct access to the report. Alternatively, you can open a report by clicking on the PRO ID listed in any page (e.g., from search or browser results).
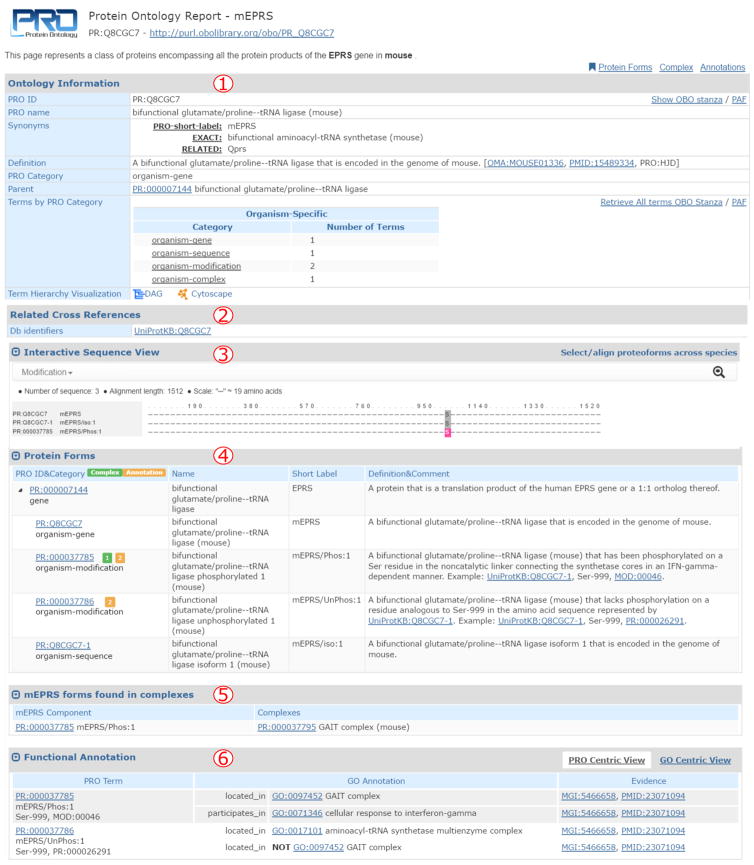
The entry report may contain some or all of the following sections (Figure 5):
Figure 5.
PRO entry report for mouse Eprs. The report shows the following sections: 1) ontology information; 2) related cross references; 3) sequence viewer; 4) protein forms; 5) subunits in complexes; and 6) annotation.
Ontology Information (Figure 5, 1): this section is found in all entries. It displays the ontological information including term ID, name, synonyms, definition, comments and parent term. A table with terms arranged by categories may be present in entries for terms that encompass many child classes (e.g., gene level, organism-gene level). This table provides a quick overview of the number of proteoforms and complexes related to a given entry, as is in the case for mouse Eprs gene in the example provided.
Related Cross References (Figure 5, 2). This section contains mappings to external databases that relate to the protein or complex report, such as UniProtKB in this example.
Interactive Sequence View (Figure 5, 3). The sequence viewer displays the protein sequence(s) defined in the entry with modified sites highlighted (color-coded based on each PTM). When the class includes more than one sequence (like our example that includes all products of mouse Eprs), a multiple sequence alignment is shown. Click on the magnifier glass to zoom in and explore specific sequence sections. The sequence viewer does not appear in protein complex reports.
Protein Forms/Complex Subunits. The Protein Forms section (found in protein reports) lists all the proteoforms related to the entry in a hierarchical way (Figure 5, 4). The Complex Subunits section (found in complex reports) lists all the proteoforms that are components of the protein complex. In our example, mouse Eprs has been observed as a phosphorylated form (on Ser-999) and in the corresponding unphosphorylated form. The numbers in the orange and green boxes next to the PRO ID indicate the presence of annotation or complex information for the particular term, respectively (see 5 and 6 below).
Forms found in complexes (found in protein reports) (Figure 5, 5). This section lists all complexes that contain at least one proteoform described on the page. For example, the Ser-999 phosphorylated form (PR:000037785) of Eprs is a component of the mouse GAIT complex (PR:000037795). The green box next to PR:000037785 in the Protein Forms table indicates that it is found in a complex.
Functional Annotation (Figure 5, 6): Finally, this section shows the annotation of the term, including functional and disease information (source: PAF file). These annotations were contributed by the PRO consortium group and by community annotators through RACE-PRO (see section 3.1.4). This table has two different views. The PRO-centric view displays the annotations for each PRO term. The annotations refer to different ontologies (e.g., GO and DO) as appropriate. On the other hand, the GO-centric view clusters all the terms that have a GO annotation in common. In that way you can see similarities among terms.
Figure 6.
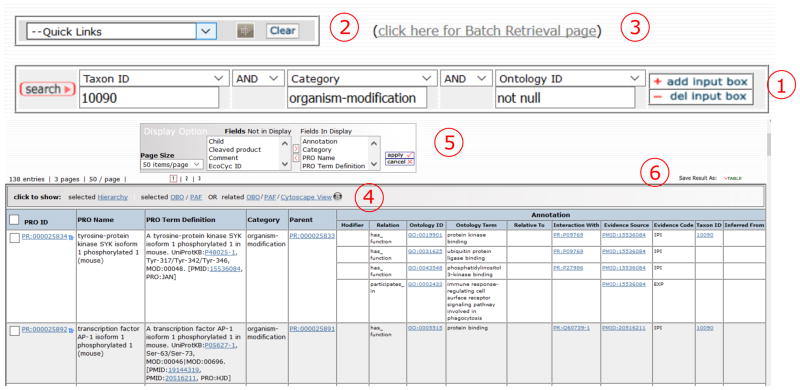
Advance search and result. 1) Search boxes with Boolean operators; 2) quick links to popular searches; 3) batch retrieval of terms using multiple identifiers; 4) result table; 5) display options to customize table content; and 6) saving option.
3.1.3. Searching PRO
Searching can be performed by entering a keyword or ID in the text Search PRO box on the right side of the homepage. For example, you can type the name of the protein for which you want to find related terms. Alternatively, the advanced search can be accessed by clicking on the Search PRO hyperlinked title above the text entry box (Figure 3, 3) on the home page. The advanced search page (Figure 6) enables searches with Boolean operators (AND, OR, NOT), as well as null (not present)/not null (present) searches with several field options (see Note 1, Figure 6, 1).
Figure 6, 1 shows an example of advanced search intended to retrieve all PRO terms for mouse proteoforms (field->Taxon ID, “10090” and field->category, “organism-modification”) containing functional annotation (field->ontology ID, “not null”). In addition, the “Quick Links” menu (Figure 6, 2) gives direct access to popular searches (like searching for phosphorylated forms) and the “Batch Retrieval” link (Figure 6, 3) allows entry of multiple identifiers (e.g., PRO and UniProt) in a single search.
In our search, 138 mouse proteoforms are shown in a results table (Figure 6, 4) with the following default columns: PRO ID, PRO name, PRO Term Definition, Category, Parent (term ID), and the searched fields. Some of the functionality in this page includes:
Display Option (Figure 6, 5): Allows you to customize the result table by adding or removing columns. Use > to add or < to remove items from the list. Click the apply button for changes to take effect.
Link to PRO entry reports: Clicking any hyperlinked PRO ID takes you to the corresponding PRO Entry report page.
Link to hierarchical view: Clicking the blue hierarchy icon next to a PRO ID opens the browser with the selected term highlighted.
Save Result As (Figure 6, 6): Allows you to save the result table as a tab-delimited file.
3.1.4. Annotation and PRO ID Requests
The annotation section is a forum for community interaction. There are two options: 1) the PRO tracker allows submission of new terms requests or changes/comments on existing ones, and 2) the rapid annotation interface, RACE-PRO, enables users to contribute directly to the curation of proteoforms.
The RACE-PRO interface can be used to:
Submit a request for a PRO ID for a proteoform of interest based on experimental evidence.
Add annotation to a proteoform or protein complex. Currently, in most databases the annotation is added to the canonical protein. There is little or no distinction made between functions of isoforms or modified forms. Using RACE-PRO, the annotation can be associated with the most appropriate protein form. Therefore, if a paper shows that only a phosphorylated form of isoform 2 of protein x is localized to the nucleus, then this annotation can be added only to PRO entry for the phosphorylated form of isoform 2, and not to others. Only experimental information is added. Another important consideration is that information submitted via RACE-PRO has to be pertinent to a particular protein sequence in a particular species.
How to Use RACE-PRO
In this section, we will demonstrate how to create the Ser-999 phosphorylated proteoform of mouse Eprs described in section 3.1.2 and shown in the Protein Forms table in the entry report in Figure 5, based on the information found in PMID:23071094. Before using RACE-PRO, the first step is to check if the proteoform is already in PRO by searching for the protein name or its UniProt accession in the search box on the home page. If it is not already in PRO, proceed to the RACE-PRO interface, as shown in Figure 7. To access the RACE-PRO interface, it is necessary to fill in minimal personal information (name, e-mail address, institution) for the purpose of saving and accessing your data and for communication; this information will not be distributed to any third party or made publically available. The save option allows you to save your information to submit at a later time. Submit is used when you are done with the entry.
Figure 7.
RACE-PRO annotation interface
Definition of the Protein Object
In this block, enter all the information about a proteoform along with the evidence source.
Retrieve the sequence: Enter a UniProt accession to retrieve the relevant sequence. For example, for mouse Eprs, enter Q8CGC7 (see Note 2). The sequence will be displayed in the box on the RACE-PRO page (see Note 4).
Specify sequence region: allows selection of a subsequence in the case of cleaved products. After saving, the selected region will be underlined.
Indicate post-translational modifications: to describe a modification, or multiple co-occurring modifications, enter the residue number and the type of modification (see Note 4). The residue number should always refer to the sequence displayed in the sequence box. After saving, the residue(s) will be highlighted (in this example residue: 999, modification: phosphorylation). Check that the highlighted residues are in the expected positions. If there is no information about any post-translational modification, then leave these field blanks. It is also possible to indicate the modifying enzyme (e.g., kinase) if such information is available.
Protein object name: add names by which this object is referred to in the paper or source of data. By default the protein name in the UniProt record is displayed. Additional synonyms can be added separated by semicolons (;).
Evidence Source: Enter information about the source of the proteoform information. In this example, select PMID from the drop-down menu and add the ID 23071094. If the appropriate option is not present in the drop-down menu, use the “Other” option. In addition, the Evidence code menu is used to select if the information is experimental, based on similarity to another proteoform or deduced by the user based on a combination of sources and knowledge.
Assay evidence: use this to indicate if the data is from in vivo or in vitro experiments.
Annotation of the Protein Object
In this block annotation from experimental data that is pertinent to the protein form described in the previous section should be added. All the information about the different columns in the table is described in the PAF guidelines. This section is optional.
What Happens Next?
An editor from the PRO team will review the entry and request any additional information if needed. The corresponding PRO term will be generated along with associated annotations (if submitted). These will have the corresponding source attribution.
3.2. Interesting Examples
3.2.1. Proteoforms with Common Annotation
14-3-3 proteins are a family of proteins that bind to phosphorylated proteins and can affect the function of the target protein in many ways including the modulation of its enzyme activity, its subcellular localization, its structure and stability, or its molecular interactions [30]. To identify proteins that are regulated by these important modulators, search for terms that are annotated with the GO term “14-3-3 protein binding” (GO:0071889). Select the search field “Annotation term” and enter “14-3-3 protein binding” in the box. Some examples are listed in Table 1. As expected all the proteins listed are phosphorylated forms, although they may contain other modifications as well. The specific 14-3-3 binding partner is listed in the “interacts with” column. However, note that the last entry, a phosphoproteoform of FOXO1 is explicitly annotated as NOT binding to 14-3-3 proteins. Therefore, this last example should be excluded in the final list of the phosphoproteoforms binding to 14-3-3 proteins.
Table 1.
Proteoforms with 14-3-3 protein binding annotation (partial results).
| PRO ID | PRO short | Relation | Ontology term (ID) | Interaction with | Evidence source |
|---|---|---|---|---|---|
| PR:000025725 | HSF1-pSer303/pSer307 | has_function | 14-3-3 protein binding (GO:0071889) | PR:P62258 | PMID:12917326 |
| PR:000025837 | MDM4-pSer367/ub | PR:P27348; PR:P61981 | PMID:16511560; PMID:16511572 | ||
| PR:000026140 | BCL2-pSer75/pSer-99 | PR:P63101 | PMID:16932738 | ||
| PR:000026848 | RFWD2-pSer387 | PR:O70456 | PMID:20843328 | ||
| PR:000029002 | HDAC4-pS210/pS246 | PMID:17179159 | |||
| PR:000029006 | HDAC5-pSer259/pSer498 | PMID:11114197 | |||
| PR:000044506 | ABL1-pThr735 | PR:000044507; PR:P63104 | PMID:15696159; PMID:16888623 | ||
| PR:000044510 | CTNNB1-pSer552 | PR:P63104 | PMID:17287208 | ||
| PR:000044814 | CSF2RB-pSer601 | PR:P63104 | PMID:10477722 | ||
| PR:000044815 | FBXO4-pSer12 | PR:P62260 | PMID:21242966 | ||
| PR:000044817 | PACS2-pSer437 | PR:000044507 | PMID:19481529 | ||
| PR:000044818 | PLK1-pSer99 | PR:P61981 | PMID:23695676 | ||
| PR:000044819 | PAK 4-pSer99/pSer474 | PR:P61981 | PMID:23695676 | ||
| PR:000027894 | FOXO1-pSer249 | NOT_has_function | 14-3-3 protein binding (GO:0071889) | PMID:18356527 |
3.2.2. Proteoform Conservation Across Species
The appetite-regulating hormone or Ghrelin is an endogenous peptidic hormone regulating both hunger and adiposity [31]. Acylated ghrelin induces a positive energy balance, while deacylated ghrelin has been reported to be devoid of any endocrine activities [32]. The hierarchy for this protein in PRO can be found at http://www.proconsortium.org/cgi-bin/pro/browser_pro?ids=PR:000007973#O and it reveals two isoforms: PR:000043841 and PR:000043842, with ortho-isoforms from human, mouse and rat as child terms. Moreover, the mouse and human active cleaved acylated forms of Ghrelin are also conserved (PR:000044483 and PR:000044484, respectively) and both are children of the organism non-specific modification term (PR:000044482).
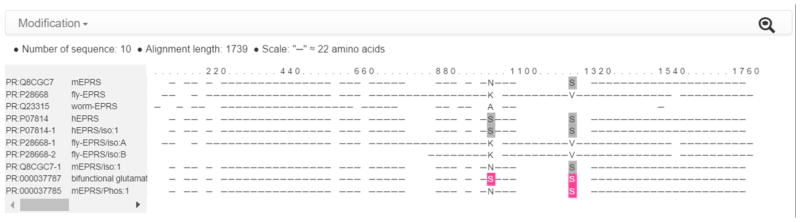
Following the previous example of the mouse Eprs, the Interactive Sequence View (Figure 5, 3) can be used to check the proteoform conservation between mouse and other species. The link on the right upper corner of the viewer expands the sequence viewer to include other related sequences in PRO. Figure 8 shows the multiple alignment, in which the combination of modified residues in each proteoform is highlighted. This particular example points to differences between human and other species: human EPRS has two serine residues that can be phosphorylated (shown in gray in PR:P07814) whereas in other species the first one is not conserved (e.g., it is asparagine in mouse).
Figure 8.
Sequence viewer for mouse Eprs proteoforms and related PRO terms. Multiple sequence alignment with highlighting of the combination of modified residues in each proteoform (pink: experimentally shown to be phosphorylated; gray: phosphorylation site conserved).
3.2.3. Find Proteoforms and Complexes Associated with Disease
PRO includes annotations of proteoforms or complexes related to disease. These annotations are connected to disease ontology terms via two relations: associated_with_disease_progression and associated_with_disease_suppression. You can retrieve all the proteins and complexes that have been annotated in the ontology with “associated_with_disease_progression”, by searching for this relation in the ontology (select the search field “Relation” and enter “association_with_disease_progression” in the box), or you could find associations for a particular disease (select the search field “Ontology term” and enter a disease name, e.g., “cancer”).
3.2.4. Comparing Protein Complexes
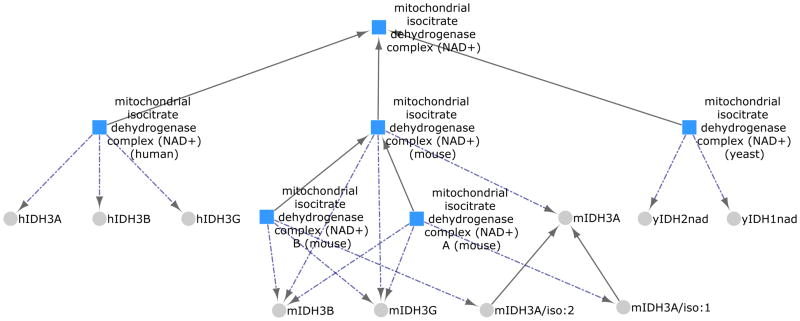
The mitochondrial isocitrate dehydrogenase complex (NAD+) catalyzes the oxidative decarboxylation of isocitrate and it is important for the regulatory control of mitochondrial energy metabolism. If you search PRO using “mitochondrial isocitrate dehydrogenase complex (NAD+)” you will retrieve all the complexes currently in PRO, including a human, a yeast and three mouse complexes. All of these species-specific complexes are children of the complex term GO:0005962 mitochondrial isocitrate dehydrogenase complex (NAD+). To review similarities and differences between the different complexes, select all the checkboxes and then click on Cytoscape link. The graphical view will open in a separate window. Figure 9 shows the complexes (squares) and their protein components (circles) connected by the relation has_component represented by the dashed arrows (see Note 5). The parent GO complex term is defined in terms of the enzymatic activity (Mitochondrial complex that possesses isocitrate dehydrogenase (NAD+) activity). The Cytoscape view of the organism-specific complexes reveals the different composition in yeast versus the human and mouse. While the yeast complex is composed of two distinct proteins, the human and mouse complexes all have three. Selecting a node provides the definition of the term and also links to its report. The relationship between the three complexes for mouse is clearly show, with one being the parent of the other two that differ with respect to which isoform of the alpha subunit is present.
Figure 9.
Cytoscape view for the mitochondrial isocitrate dehydrogenase complex (NAD+) in PRO. Complexes are represented by squares, proteins by circles. Solid arrows represent the is_a relation, while broken arrows represent the has_component relation.
Another rich example is provided by complexes of cyclin dependent kinases (CDKs) with cyclins. CDKs are a family of multifunctional enzymes that can modify various protein substrates involved in cell cycle progression. Their activity is controlled by their phosphorylation state and their binding to a cyclin regulatory subunit. All CDK:cyclin complexes in PRO are under cyclin-dependent protein kinase holoenzyme complex (GO:0000307). The hierarchical view of this term in PRO (http://www.proconsortium.org/cgi-bin/pro/browser_pro?ids=GO:0000307) lists all the complexes of CDKs and cyclins that have been curated. The Cytoscape view can provide a more granular level where the specific subunits, including the modifications, can be displayed. For example, PRO contains five different “cyclin B1:cdk1 complex” for human which differ in the phosphorylation state of its component. Thus, the Cytoscape view in PRO facilitates the comparison of complexes within and across species.
Acknowledgments
PRO Consortium participants: Protein Information Resource, The Jackson Laboratory, Reactome, and the New York State Center of Excellence in Bioinformatics and Life Sciences. PRO is funded by NIH grant R01GM080646.
Footnotes
- To retrieve all the entries from a given category, for example, all the nodes for gene product level, search by selecting the field “category” and entering “gene” in the box.
- Some of the search fields are of the type null/not null. This is the case for the ortho-isoform and ortho-modified form. To retrieve the ortho-isoform entries, select the search field ortho-isoform and type not null.
- More details about the options for the DB ID, Modifiers, and Relations fields are listed in the PAF guidelines (see Materials).
To search UniProt accessions at the UniProt website (www.uniprot.org), enter the protein name and organism: eprs and mouse into the search box. From the result list, check the one that is relevant to your search. Alternatively, enter eprs and then use the filter to select mouse as the organism. View likely UniProt entries to confirm that it represents the protein of interest. If a published paper describes a particular isoform, also check if this isoform is already present in UniProt (in the Sequences section). You can also enter UniProt identifiers for isoforms (a UniProtKB accession followed by a dash and a number, e.g., Q8CGC7-1). If you have an identifier from a different database, use UniProt’s ID mapping service (http://www.uniprot.org/uploadlists/) to obtain the corresponding UniProtKB accession and retrieve the sequence.
The UniProt sequence retrieved is formatted to show the residue numbers, and the organism box is automatically filled in.
If the modification is not in the list, use the “Other” option to add it. These terms will be later mapped to the corresponding PSI-MOD terms (e.g., Ser phosphorylation will become MOD:00046). If the modification site is unknown, please enter “?” in the residue number box. Enter one modification site on each line. Use the [more] or [less] buttons to add or remove a modification line.
The Cytoscape view may be complex, and you may want to hide nodes to focus on a specific part of the graph. To hide all the protein nodes, select “display option” from the top menu bar and uncheck “All Protein” under “Nodes Type”. Otherwise, selecting any node or set of nodes and right clicking will open a menu with the option to hide nodes.
References
- 1.Shihab HA, Gough J, Mort M, Cooper DN, Day IN, Gaunt TR. Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Hum Genomics. 2014;8:11. doi: 10.1186/1479-7364-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omenn GS, Yocum AK, Menon R. Alternative splice variants, a new class of protein cancer biomarker candidates: findings in pancreatic cancer and breast cancer with systems biology implications. Dis Markers. 2010;28(4):241–251. doi: 10.3233/dma-2010-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menon R, Zhang Q, Zhang Y, Fermin D, Bardeesy N, DePinho RA, Lu C, Hanash SM, Omenn GS, States DJ. Identification of novel alternative splice isoforms of circulating proteins in a mouse model of human pancreatic cancer. Cancer Res. 2009;69(1):300–309. doi: 10.1158/0008-5472.can-08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mair W, Muntel J, Tepper K, Tang S, Biernat J, Seeley WW, Kosik KS, Mandelkow E, Steen H, Steen JA. FLEXITau: Quantifying Post-translational Modifications of Tau Protein in Vitro and in Human Disease. Anal Chem. 2016;88(7):3704–3714. doi: 10.1021/acs.analchem.5b04509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kessler BM. Ubiquitin - omics reveals novel networks and associations with human disease. Curr Opin Chem Biol. 2013;17(1):59–65. doi: 10.1016/j.cbpa.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 7.Jin K, Musso G, Vlasblom J, Jessulat M, Deineko V, Negroni J, Mosca R, Malty R, Nguyen-Tran DH, Aoki H, Minic Z, Freywald T, Phanse S, Xiang Q, Freywald A, Aloy P, Zhang Z, Babu M. Yeast mitochondrial protein-protein interactions reveal diverse complexes and disease-relevant functional relationships. J Proteome Res. 2015;14(2):1220–1237. doi: 10.1021/pr501148q. [DOI] [PubMed] [Google Scholar]
- 8.Climer LK, Dobretsov M, Lupashin V. Defects in the COG complex and COG-related trafficking regulators affect neuronal Golgi function. Front Neurosci. 2015;9:405. doi: 10.3389/fnins.2015.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Natale DA, Arighi CN, Blake JA, Bult CJ, Christie KR, Cowart J, D’Eustachio P, Diehl AD, Drabkin HJ, Helfer O, Huang H, Masci AM, Ren J, Roberts NV, Ross K, Ruttenberg A, Shamovsky V, Smith B, Yerramalla MS, Zhang J, AlJanahi A, Celen I, Gan C, Lv M, Schuster-Lezell E, Wu CH. Protein Ontology: a controlled structured network of protein entities. Nucleic Acids Res. 2014;42(Database issue):21. doi: 10.1093/nar/gkt1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith LM, Kelleher NL. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10(3):186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arighi C, Shamovsky V, Masci AM, Ruttenberg A, Smith B, Natale DA, Wu C, D’Eustachio P. Toll-like receptor signaling in vertebrates: testing the integration of protein, complex, and pathway data in the protein ontology framework. PLoS One. 2015;10(3):e0122978. doi: 10.1371/journal.pone.0122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bult CJ, Drabkin HJ, Evsikov A, Natale D, Arighi C, Roberts N, Ruttenberg A, D’Eustachio P, Smith B, Blake JA, Wu C. The representation of protein complexes in the Protein Ontology (PRO) BMC Bioinformatics. 2011;12:371. doi: 10.1186/1471-2105-12-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altenhoff AM, Skunca N, Glover N, Train CM, Sueki A, Pilizota I, Gori K, Tomiczek B, Muller S, Redestig H, Gonnet GH, Dessimoz C. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015;43(Database issue):D240–249. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Celen I, Ross KE, Arighi CN, Wu CH. Bioinformatics Knowledge Map for Analysis of Beta-Catenin Function in Cancer. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0141773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):26. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montecchi-Palazzi L, Beavis R, Binz PA, Chalkley RJ, Cottrell J, Creasy D, Shofstahl J, Seymour SL, Garavelli JS. The PSI-MOD community standard for representation of protein modification data. Nat Biotechnol. 2008;26(8):864–866. doi: 10.1038/nbt0808-864. [DOI] [PubMed] [Google Scholar]
- 18.Mungall CJ, Batchelor C, Eilbeck K. Evolution of the Sequence Ontology terms and relationships. J Biomed Inform. 2011;44(1):87–93. doi: 10.1016/j.jbi.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium U. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42(Database issue):191–198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meldal BH, Forner-Martinez O, Costanzo MC, Dana J, Demeter J, Dumousseau M, Dwight SS, Gaulton A, Licata L, Melidoni AN, Ricard-Blum S, Roechert B, Skyzypek MS, Tiwari M, Velankar S, Wong ED, Hermjakob H, Orchard S. The complex portal--an encyclopaedia of macromolecular complexes. Nucleic Acids Res. 2015;43(Database issue):D479–484. doi: 10.1093/nar/gku975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y, Xiang Z, He Y. Ontology-based representation and analysis of host-Brucella interactions. J Biomed Semantics. 2015;6:37. doi: 10.1186/s13326-015-0036-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eppig JT, Richardson JE, Kadin JA, Ringwald M, Blake JA, Bult CJ. Mouse Genome Informatics (MGI): reflecting on 25 years. Mamm Genome. 2015;26(7–8):272–284. doi: 10.1007/s00335-015-9589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDowall MD, Harris MA, Lock A, Rutherford K, Staines DM, Bahler J, Kersey PJ, Oliver SG, Wood V. PomBase 2015: updates to the fission yeast database. Nucleic Acids Res. 2015;43(Database issue):D656–661. doi: 10.1093/nar/gku1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen M, Cox AP, Chaudhry N, Ng M, Sule D, Duncan W, Ray P, Weinstock-Guttman B, Smith B, Ruttenberg A, Szigeti K, Diehl AD. The neurological disease ontology. J Biomed Semantics. 2013;4(1):42. doi: 10.1186/2041-1480-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bada M, Eckert M, Evans D, Garcia K, Shipley K, Sitnikov D, Baumgartner WA, Jr, Cohen KB, Verspoor K, Blake JA, Hunter LE. Concept annotation in the CRAFT corpus. BMC Bioinformatics. 2012;13:161. doi: 10.1186/1471-2105-13-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham F, Moore B, Ruiz-Schultz N, Ritchie GR, Eilbeck K. Improving the Sequence Ontology terminology for genomic variant annotation. J Biomed Semantics. 2015;6:32. doi: 10.1186/s13326-015-0030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kibbe WA, Arze C, Felix V, Mitraka E, Bolton E, Fu G, Mungall CJ, Binder JX, Malone J, Vasant D, Parkinson H, Schriml LM. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015;43(Database issue):27. doi: 10.1093/nar/gku1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obsilova V, Kopecka M, Kosek D, Kacirova M, Kylarova S, Rezabkova L, Obsil T. Mechanisms of the 14-3-3 protein function: regulation of protein function through conformational modulation. Physiol Res. 2014;63(Suppl 1):S155–164. doi: 10.33549/physiolres.932659. [DOI] [PubMed] [Google Scholar]
- 31.Nogueiras R, Williams LM, Dieguez C. Ghrelin: new molecular pathways modulating appetite and adiposity. Obesity facts. 2010;3(5):285–292. doi: 10.1159/000321265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asakawa A, Inui A, Fujimiya M, Sakamaki R, Shinfuku N, Ueta Y, Meguid MM, Kasuga M. Stomach regulates energy balance via acylated ghrelin and desacyl ghrelin. Gut. 2005;54(1):18–24. doi: 10.1136/gut.2004.038737. [DOI] [PMC free article] [PubMed] [Google Scholar]