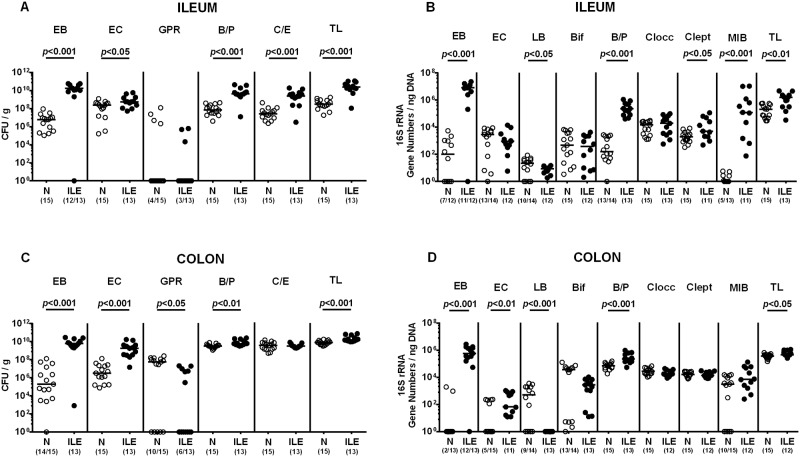
Fig 1. Intestinal microbiota composition in human microbiota associated mice suffering from acute ileitis.
Human microbiota associated (hma) were perorally infected with T. gondii strain ME49 to induce acute ileitis as described in methods. Main intestinal bacterial groups abundant in the ileum (A, B) and the colon lumen (C, D) of hma mice were quantitatively assessed applying both culture (A, C) and culture-independent (i.e. molecular 16S rRNA based; B, D) methods 7 days following ileitis induction (ILE, filled circles). Noninfected hma mice served as controls (N, open circles). Numbers of enterobacteria (EB), enterococci (EC), Gram-positive rods (GPR), Bacteroides / Prevotella spp. (B/P), Clostridium / Eubacterium spp. (C/E) and the total bacterial loads (TL) are expressed as colony forming units per gram feces (CFU / g). 16S rRNA of the main intestinal bacterial groups including enterobacteria (EB), enterococci (EC), lactobacilli (LB), bifidobacteria (Bif), Bacteroides / Prevotella species (B/P), Clostridium coccoides group (Clocc), Clostridium leptum group (Clept), Mouse Intestinal Bacteroides (MIB) and the total eubacterial loads (TL) are expressed as gene numbers per ng DNA. Numbers of animals harboring the respective bacterial group out of the total number of analyzed mice are given in parentheses. Medians (black bars) and significance levels (p-values) determined by Student’s t test and Mann-Whitney U test are indicated. Data shown are pooled from three independent experiments.