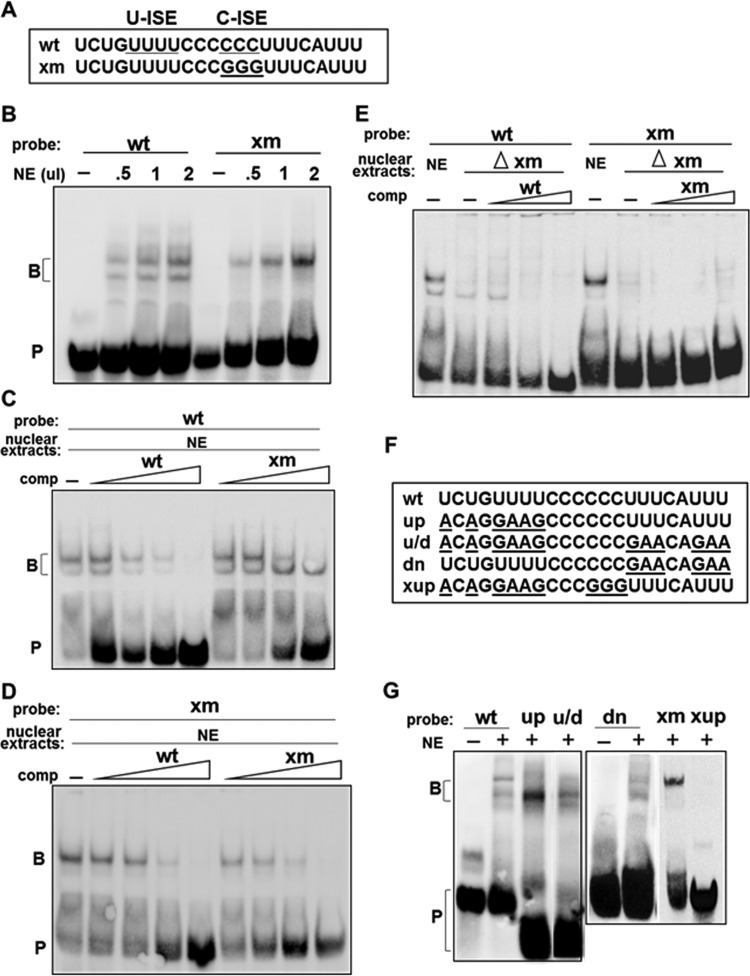
FIG 2.
Two distinct RNA-protein complexes are formed on the U-ISE and C-ISE regions. (A) Wild type (wt) and C-mutated (xm) RNA probes used for EMSA. (B) wt probe formed two RNA-protein complexes; xm probe formed one complex in HeLa extracts. NE, nuclear extract (in microliters); B, RNA-protein complex; P, probe. (C and D) Specificity of the RNA-protein complexes analyzed by competition assay. A 2- to 50-fold molar excess of unlabeled wt or xm RNAs (comp, competitor) was added along with the biotinylated wt or xm probe. −, probe incubated without competitors. (E) HeLa cell nuclear extract lacking xm binding proteins (Δxm) do not form upper RNA complexes. Extracts were depleted by preincubation with streptavidin-Sepharose beads containing biotinylated xm probes. Depleted extracts were analyzed by EMSA with wt or xm probe in the absence (−) or presence of a 5-, 20-, or 50-fold molar excess of unlabeled competitors (comp). (F) Probes used to map the RNA-protein complex binding sites shown in panel G. (G) EMSA performed using the wt or mutated probe (up, u/d, dn, xm, and xup) and HeLa nuclear extracts. −, probe alone; B, RNA-protein complex; P, probe.