ABSTRACT
Only 1% of marine bacteria are currently culturable using standard laboratory procedures, and this is a major obstacle for our understanding of the biology of marine microorganisms and for the discovery of novel microbial natural products. Therefore, the purpose of this study was to investigate if improved cultivation conditions, including the use of an alternative gelling agent and supplementation with signaling molecules, improve the culturability of bacteria from seawater. Replacing agar with gellan gum improved viable counts 3- to 40-fold, depending on medium composition and incubation conditions, with a maximum of 6.6% culturability relative to direct cell counts. Through V4 amplicon sequencing we found that culturable diversity was also affected by a change in gelling agent, facilitating the growth of orders not culturable on agar-based substrates. Community analyses showed that communities grown on gellan gum substrates were significantly different from communities grown on agar and that they covered a larger fraction of the seawater community. Other factors, such as incubation temperature and time, had less obvious effects on viable counts and culturable diversity. Supplementation with acylated homoserine lactones (AHLs) did not have a positive effect on total viable counts or a strong effect on culturable diversity. However, low concentrations of AHLs increased the relative abundance of sphingobacteria. Hence, with alternative growth substrates, it is possible to significantly increase the number and diversity of cultured marine bacteria.
IMPORTANCE Serious challenges to human health, such as the occurrence and spread of antibiotic resistance and an aging human population in need of bioactive pharmaceuticals, have revitalized the search for natural microbial products. The marine environment, representing the largest ecosystem in the biosphere, harbors an immense and virtually untapped microbial diversity producing unique bioactive compounds. However, we are currently able to cultivate only a minute fraction of this diversity. The lack of cultivated microbes is hampering not only bioprospecting efforts but also our general understanding of marine microbes. In this study, we present a means to increase the number and diversity of cultured bacteria from seawater, showing that relatively simple changes to medium components may facilitate the isolation and growth of hitherto unknown bacterial orders.
KEYWORDS: AHLs, cultivation, gellan gum, marine microbiology, microbial communities
INTRODUCTION
The ocean is the largest ecosystem on Earth, covering 70% of the surface of the planet and representing the greatest pool of microbial life on the planet (1). Microorganisms are key players in the biogeochemical cycles, and their remineralization of organic matter forms the basis of all life. Furthermore, microorganisms are excellent sources of bioactive compounds, such as antibiotics, hydrolytic enzymes, and anticancer agents (2, 3). For decades, terrestrial microorganisms have been used as sources of such bioactive compounds. However, the growing need for new drugs and bioprocessing compounds has led to the expansion of bioprospecting efforts to include other environments. The marine environment, particularly marine microorganisms, is considered a very promising source of novel bioactivities. Marine microorganisms have evolved to survive and grow under conditions differing from those of terrestrial environments (e.g., high salinity, pressure, and oligotrophy). Therefore, the chemistry of many of their bioactive compounds is different from those produced by terrestrial organisms, potentially having novel targets and mechanisms of action (4–8).
The vast majority of marine microorganisms (90 to 99%) have not yet been successfully cultured under laboratory conditions (3), a phenomenon that has been referred to as the “great plate count anomaly” (9). Furthermore, cultured microorganisms are not representative of the actual diversity. For instance, cultivation-independent approaches have revealed that Alphaproteobacteria represent one of the most abundant marine bacterial classes (10, 11), while most cultured microorganisms belong to Gammaproteobacteria. Genomic and metagenomic sequencing has enabled the study of the overall function and activity of environmental microbial communities (12–17) and has also allowed the identification of novel bacterial groups and the assessment of the biotechnological potential of microbial communities by identifying biosynthetic pathways of pharmaceutical interest, and bioprocessing. However, the exploitation of metagenomes through heterologous expression and the production of fully functional products is difficult (18, 19). Therefore, there is great interest in improving the culturability of microorganisms. Furthermore, cultivation-based tools are still of paramount importance for a comprehensive understanding of the biology, ecology, and bioactivity of prokaryotes and are essential for future bioprospecting (1, 19).
For the last 2 decades, scientists have tried to bring “not-yet-culturable” species into culture, and a range of approaches have indeed either improved overall culturability or enabled culturing of specific organisms. Typically, these approaches attempt to mimic the natural niche by reducing nutrients, adding specific compounds, or changing incubation atmosphere, temperature, and time (e.g., see references 20–25). Also, the requirements for a mixed microbial community and signals from this community have been proposed as an explanation for the lack of culturability (1, 26). Indeed, D'Onofrio et al. (27) used coculturing experiments to show that some marine microbes only grow when paired with a specific combination of other species. Similarly, a coculture approach brought between 30 and 40% of soil and seawater bacteria into culture (24).
Agar is by far the most widely used gelling agent in microbiology; however, a number of other compounds, such as xanthan gum, carrageenan, and gellan gum, have similar gelling properties. Several studies have demonstrated that solidifying growth substrates with gellan gum, which is a fermentation product of Sphingomonas elodea (28), appears to enhance the culturability of some microorganisms. Gellan gum solidifies at room temperature in the presence of multivalent cations such as magnesium and calcium (28, 29), and its high thermal stability makes it ideal for the isolation and growth of several thermophilic microorganisms (30–33). Studies on soil and freshwater sediment bacteria have shown that alternative gelling agents, such as gellan gum, may enable more microorganisms to grow and form colonies on solid substrates than on substrates solidified with agar (30–32, 34–37), resulting in the isolation of previously unculturable bacteria (35, 38). Improving the culturability of marine bacteria through the application of alternative gelling agents in growth substrates has so far not been done. Hence, the purpose of the present study was to determine how gellan gum, in comparison with agar, affects the overall culturability and the culturable diversity of microbial communities from seawater. Furthermore, the effect of specific quorum-sensing signaling molecules on culturability was assessed.
RESULTS
Coastal surface seawater was sampled on two occasions, sampling 1 (S1) and sampling 2 (S2). Microbial abundances were determined by direct cell counts and by colony counts on different growth substrates based on either a nutrient-rich medium (marine broth; MB) or a nutrient-poor medium (filtered seawater; SW) solidified with either agar (MBA and SWA, respectively) or gellan gum (MBG and SWG, respectively). Plates were incubated at 10°C for 5 or 10 weeks or at 25°C for 2 or 5 weeks. In S2, some SWA and MBA plates were further supplemented with two different concentrations of acylated homoserine lactones (AHLs). Viable counts were determined and the compositions and diversity of the culturable communities were assessed and compared with those from seawater communities by V4 sequencing.
In situ abundances of planktonic bacteria, as estimated by SYBR gold staining and fluorescence microscopy, were 8.4 × 105 cells ml−1 ± 2.3 × 105 cells ml−1 and 5.8 × 106 cells ml−1 ± 1.6 × 106 cells ml−1 for S1 and S2, respectively. The viable counts (CFU) on solid growth media ranged from 70 ± 37 CFU ml−1 for seawater agar (SWA) incubated at 10°C for 5 weeks (S1) to 3.8 × 105 ± 1.4 × 105 CFU ml−1 for marine broth gellan gum (MBG) incubated at 25°C for 5 weeks (S2). Generally, MBG enhanced the culturability compared with that of the other substrates, with 2.4 to 6.6% of bacteria being culturable on this substrate depending on sampling time point and incubation conditions (Table 1). The equivalent medium solidified with agar (MBA) exhibited significantly less culturability with 0.4 to 2.3% (P = 0.005). The less nutrient-rich seawater media showed lower culturability than the richer marine broth-based media, and on these substrates, gelling agent had an even more marked impact on culturability (P < 0.001).
TABLE 1.
Culturability of bacteria
| Substrate | Culturability (%)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| S1 |
S2 |
|||||||
| 10°C |
25°C |
10°C |
25°C |
|||||
| 5 wk | 10 wk | 2 wk | 5 wk | 5 wk | 10 wk | 2 wk | 5 wk | |
| MBA | 0.4 | 0.7 | 1.5 | 1.5 | 0.4 | 0.4 | 0.7 | 2.3 |
| MBAAHL0.5 | 0.2 | 0.3 | 1.3 | 1.8 | 0.1 | 0.2 | 0.6 | 1.4 |
| MBAAHL50 | 0.1 | 0.1 | 0.2 | 0.2 | 0.1 | 0.2 | 0.1 | 0.1 |
| MBG | 2.5 | 2.7 | 3.8 | 4.1 | 3.2 | 3.4 | 2.4 | 6.6 |
| SWA | 0.01 | 0.03 | 0.02 | 0.02 | 0.01 | 0.02 | 0.003 | 0.005 |
| SWAAHL0.5 | 0.01 | 0.02 | 0.01 | 0.01 | 0.01 | 0.02 | 0.007 | 0.01 |
| SWAAHL50 | 0.01 | 0.01 | 0.01 | 0.03 | 0.002 | 0.004 | 0.002 | 0.003 |
| SWG | 0.1 | 0.2 | 0.4 | 0.6 | 0.2 | 0.2 | 0.1 | 0.2 |
Given as CFU (CFU ml−1) relative to total cell counts (cells ml−1) in seawater.
Microbial community composition and culturable diversity.
From S1 and S2 combined, a total of 1.4 × 107 V4 amplicon sequences were retained after cleaning. Clustering the sequences at 97% nucleotide sequence similarity produced 7,647 and 7,594 operational taxonomic units (OTUs) for S1 and S2, respectively. The numbers of sequences per sample varied between 3.8 × 103 and 6.8 × 105 (see Table S1 in the supplemental material) with a mean and standard deviation of 1.3 × 105 ± 9.7 × 104.
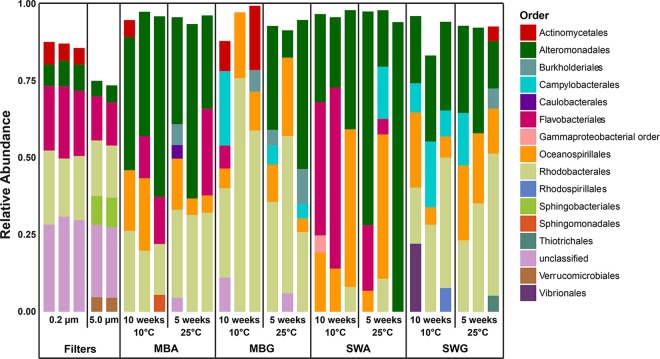
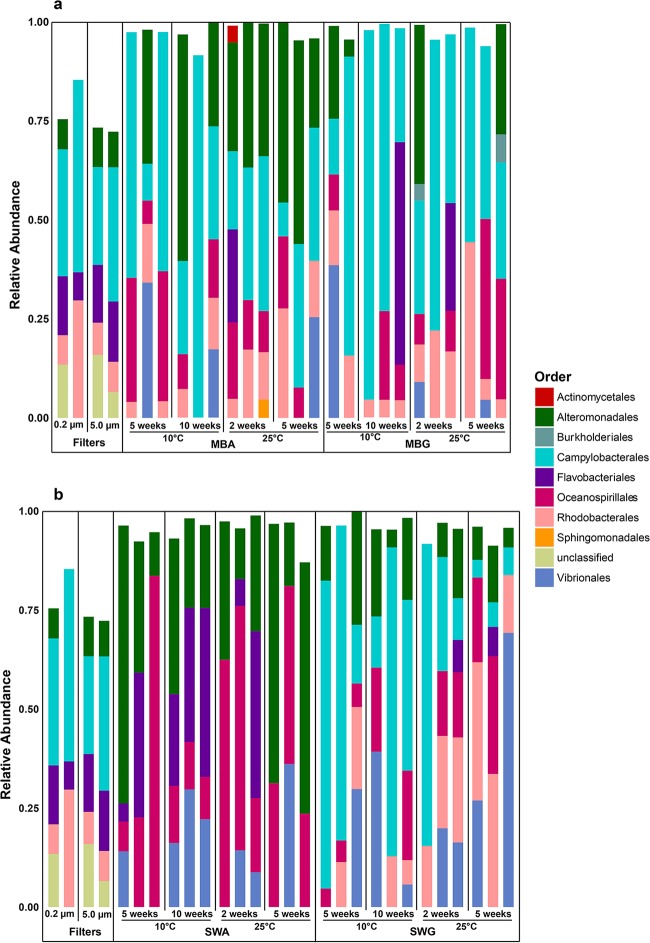
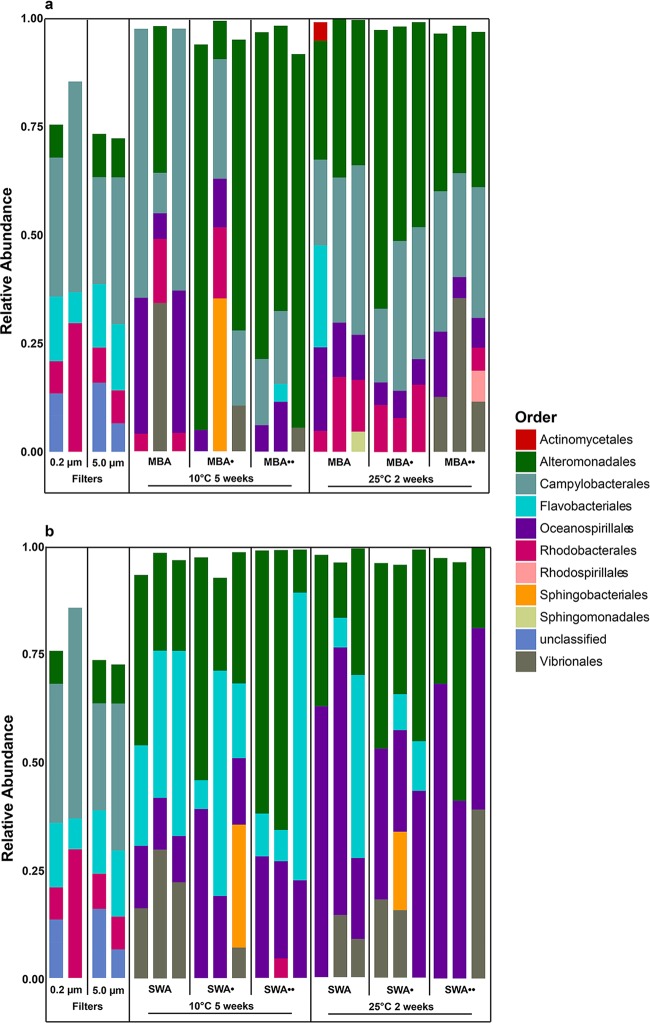
At the time of S1, the particle-associated microbial community (≥5 μm) in the seawater was dominated by Alteromonadales, Flavobacteriales, Rhodobacterales, Sphingobacteriales, Verrucomicrobiales, and a large fraction of bacteria unclassified at the order level (Fig. 1). Similarly, Alteromonadales, Flavobacteriales, Rhodobacterales, and unclassified orders dominated the free-living, planktonic fraction (≥0.2 μm to <5 μm) but the Sphingobacteriales and Verrucomicrobiales decreased in relative abundance, whereas the Actinomycetales increased in abundance relative to the particle-associated communities. Low-abundance (<4%) OTUs accounted for approximately 25% of the sequences in the large-size fraction, and the majority of the microbial diversity present in the seawater at this time point was, according to Chao1 richness estimates and Shannon diversity index values (see Fig. S2 in the supplemental material), associated with particles. Approximately 1 month later, during S2, a different microbial community, dominated by Campylobacterales (approximately 25 to 50%), was present in both size fractions (Fig. 2). Concomitantly, measures of alpha-diversity were lower during S2 than during S1 (Fig. S2) and in contrast to S1, the planktonic fraction of the microbial community was more diverse than the particle-associated fraction during S2.
FIG 1.
Culturable community compositions during sampling 1 compared with that of the in situ community. The compositions are depicted as relative proportions of bacterial orders obtained from seawater (5 μm, 0.2-μm filters) and from cultivated bacteria on marine broth agar (MBA), marine broth gellan gum (MBG), seawater agar (SWA), and seawater gellan gum (SWG).
FIG 2.
Culturable community compositions during sampling 2 compared with that of the in situ community. The compositions are depicted as relative proportions of bacterial orders obtained from seawater (5 μm, 0.2-μm filters) and from cultivated bacteria on marine broth agar (MBA) and marine broth gellan gum (MBG) (A) and on seawater agar (SWA) and seawater gellan gum (SWG) (B).
As for overall culturability, the compositions of the cultured fractions of the microbial communities were affected by media and gelling agents. In S1, the orders Rhodospirillales, Thiotrichales, and Vibrionales were only observed above the threshold of 4% on substrates solidified with gellan gum (Fig. 1). Two replicate marine broth plates solidified with gellan gum (MBG) exhibited high relative abundances of sequences relating to Actinomycetales (10 to 20%) in S1. These plates were incubated at 10°C for 10 weeks. This order was also represented on one seawater plate solidified with gellan gum (SWG) and one marine broth plate solidified with agar, though in lower relative abundance (Fig. 1). This was not just due to a single OTU reaching a high relative abundance on these plates, as the Actinomycetales order had a high relative OTU richness on this substrate (see Fig. S3). Campylobacterales, Burkholderiales, and Rhodobacterales also seemed to have consistently higher relative abundances on substrates solidified with gellan gum during both samplings (Fig. 1 and 2). Agar-based substrates supported the growth of Flavobacteriales better than substrates solidified with gellan gum during S1 (Fig. 1). In S2, this trend was not as obvious and most culturable communities were dominated by sequences related to the Campylobacterales order, except for the SWA plates (Fig. 2). SWA favored the growth of the Flavobacteriales and the gammaproteobacterial orders Alteromonadales and Oceanospirillales in S1, whereas the alphaproteobacterial orders were generally absent or low in abundance on these substrates (Fig. 1). Concomitantly, SWA plates also exhibited the lowest Shannon diversity values of the S1 data set (Fig. S2). Similarly, Flavobacteriales and the three gammaproteobacterial orders Alteromonadales, Oceanospirillales, and Vibrionales accounted for almost all of the sequences on these substrates in S2 (Fig. 2). Some orders, e.g., the Rhodobacterales, Campylobacterales, Oceanospirillales, Vibrionales, Alteromonadales, Actinomycetales, and Flavobacteriales, exhibited relatively high OTU richness across all substrate types and incubation conditions (see Fig. S3 and S4), whereas their relative abundances varied substantially between treatments (Fig. 1 and 2). From the community analysis, it seemed that the consistency between replicates was highest in natural communities, while in some cases, there were larger deviations in cultured communities (e.g., SWA communities in S1) (Fig. 1). This notion was substantiated by the fact that only a limited number of OTUs was shared by all three replicates in many of the treatments (see Fig. S5 to S7).
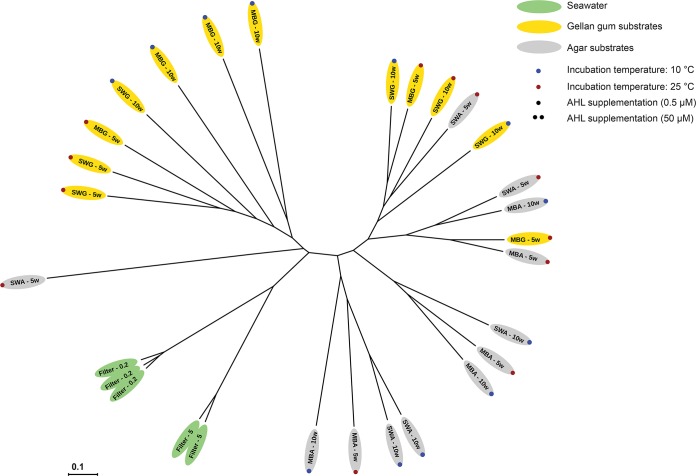
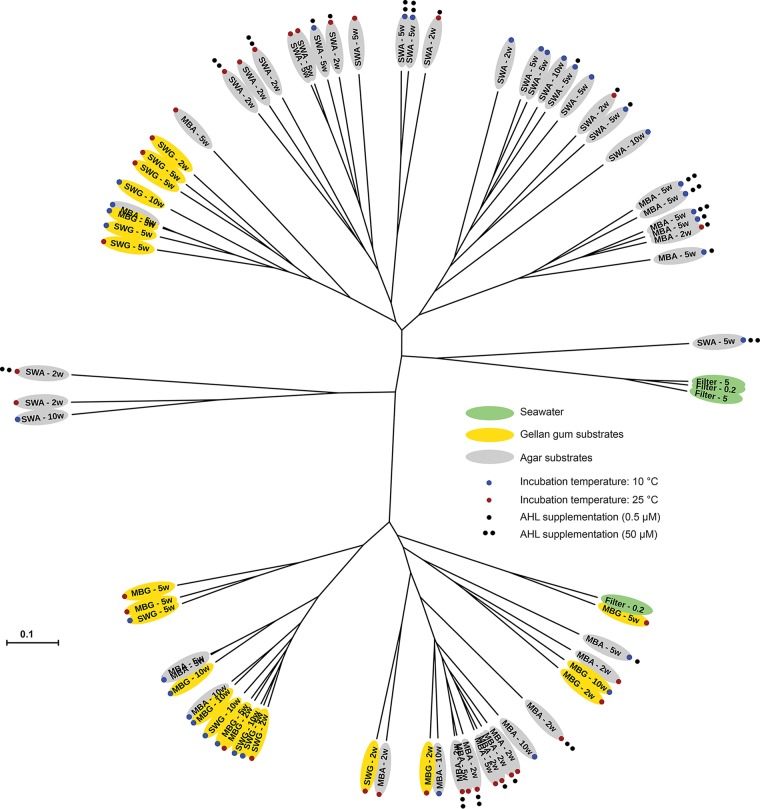
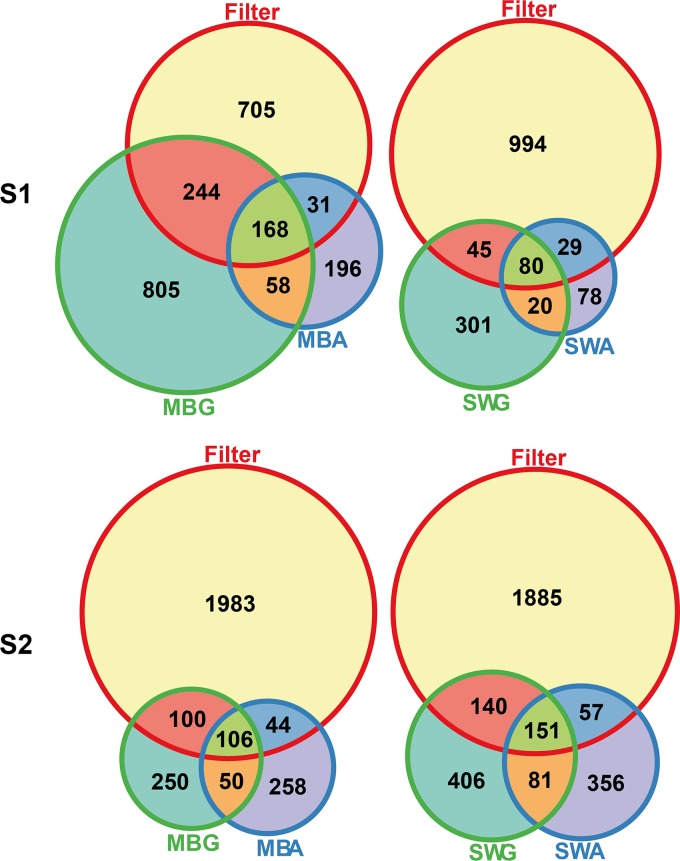
The community structure analyses substantiated the impact of gelling agent on the composition of the culturable microbial communities. From S1, the communities growing on substrates solidified with gellan gum clustered together in two overall clusters (Fig. 3). The exceptions were one community from an MBG plate that clustered with three communities from agar plates and one SWA plate incubated at 25°C for 5 weeks that clustered with one of the gellan gum subclusters. Another replicate from the same treatment did not cluster with other communities. The remaining communities growing on agar-based substrates formed three subclusters. The observed subclusters did not seem to be defined by the media or the incubation conditions (Fig. 3). A parsimony test on the UniFrac distances confirmed that the observed differences in community structure on substrates solidified with either gellan gum or agar were significant (P = 0.003). This was not the case for any of the other treatments, i.e., medium, incubation time, and incubation temperature (P > 0.05) (Table 2). In the OTU-based analysis of community structure, the same results were obtained using analyses of molecular variance (AMOVAs) on Bray-Curtis distances (Table 2). For S2, the microbial communities clustered together based both on gelling agent and on medium type (Fig. 4). The 23 gellan gum plate communities clustered together in two overall subclusters except for five plates, which spread among MBA plates or with one of the seawater communities. A total of five MBA plates also clustered with the two gellan gum subclusters, while all SWA communities clustered together in two distinct sub-subclusters. The fact that both the gelling agent and the medium influenced the community structure was substantiated by the parsimony testing and AMOVA (P ≤ 0.002) (Table 2). The effect of incubation time on the structures of the cultured microbial communities was not statistically significant according to the same analyses (P > 0.05). UniFrac data suggested that incubation temperature did have an effect (parsimony, P = 0.03), whereas this effect was not statistically significant according to the OTU-based approach (AMOVA, P = 0.07). The seawater communities were similar to each other and distinct from the cultured communities in both samplings except from one replicate in S2 that was more similar to the community from an MBG plate (Fig. 4). Whether some substrates supported the growth of a community more similar to the one present in situ was unclear on the basis of the community structure analyses, but a comparison of the numbers of seawater OTUs that were recovered on the different substrates suggested that substrates solidified with gellan gum supported the growth of more OTUs than the corresponding agar-solidified substrate, and they supported the growth of more of the OTUs observed in seawater (Fig. 5). Furthermore, this analysis also suggested that the overlap between OTUs recovered on substrates solidified with gellan gum and substrates solidified with agar was relatively small.
FIG 3.
Unrooted tree depicting weighted UniFrac distances (indicated by the scale bar) between individual samples from sampling 1 (S1). Sample names are given according to growth substrate and incubation time, except for seawater samples, which are named according to filter size (μm).
TABLE 2.
Treatment effects on community structure for S1 and S2
| Treatment | S1 P valuea |
S2 P valuea |
||
|---|---|---|---|---|
| Parsimony | AMOVA | Parsimony | AMOVA | |
| Gelling agent | 0.003 | <0.001 | 0.002 | <0.001 |
| Medium | 0.6 | 0.07 | <0.001 | <0.001 |
| Incubation time | 0.5b | 0.6b | 0.6–0.9c | 0.07–0.09c |
| Incubation temp | 0.5b | 0.6b | 0.03 | 0.07 |
| AHLs (0.5 μM) | NAd | NA | 0.08 | 0.03 |
| AHLs (50 μM) | NA | NA | 0.07 | 0.003 |
Parsimony tests were performed on weighted UniFrac distances, and analyses of molecular variance (AMOVA) were performed on Bray-Curtis distances.
Time and temperature comparisons were identical, as all plates incubated at 10°C were incubated for 10 weeks and all plates incubated at 25°C were incubated for 5 weeks.
Two different incubation times were applied for each temperature (10°C for 5 weeks and 10 weeks; 25°C for 2 weeks and 5 weeks).
NA, not applicable.
FIG 4.
Unrooted tree depicting weighted UniFrac distances (indicated by the scale bar) between individual samples from sampling 2 (S2). Sample names are given according to growth substrate and incubation time, except for seawater samples, which are named according to filter size (μm).
FIG 5.
Venn diagrams comparing the recoveries of planktonic (≥0.2 μm to <5 μm) seawater OTUs (Filter) on different substrates and showing the overlap of OTUs recovered on equivalent media solidified with either gellan gum (MBG and SWG) or agar (MBA and SWA) from sampling 1 (S1) and sampling 2 (S2). Communities from all incubation conditions were pooled.
Recovery of unclassified OTUs.
Sequences unclassified at the order level were recovered most frequently during S1 where the in situ abundance of unclassified OTUs was highest (Fig. 1 and 2). One MBA plate incubated at 25°C for 5 weeks supported the growth of unclassified OTUs, exhibiting relative abundances of approximately 5% (Fig. 1). Two MBG plates, one incubated at 10°C for 10 weeks, and one incubated at 25°C for 5 weeks, also exhibited relatively high abundances of unclassified OTUs with ca. 5 and 10%, respectively (Fig. 1). One OTU, OTU279, represented 5,798 sequences in S1, and 5,360 (92%) of these were derived from MBG plates. BLAST results showed that this OTU exhibited 100% nucleotide sequence similarities to numerous environmental sequences from various marine environments (e.g., accession no. KR077719, KR077314, KT731894, and KM224111). It also exhibited 95 to 100% similarities to six uncultured putative actinobacteria isolated from coastal water using fluorescence-activated cell sorting (accession no. JF488172, HQ675198, HQ675197, HQ675140, HQ675191, and HQ675143) and 95% nucleotide sequence similarity to an uncultured member of a proposed novel marine actinobacterial subclass, the “Candidatus Actinomarinidae” known only from metagenomes (39). Its closest described relative, the actinobacterium Conexibacter woesei, showed 81% nucleotide sequence similarity to OTU279. Similar observations were found for abundant unclassified beta- and gammaproteobacterial OTUs.
Impact of AHLs on culturability.
Supplementation with AHLs had a negative or no effect on viable counts (Table 1). It did affect microbial community composition in some instances during S2 (Fig. 6). For MBA substrates incubated at 10°C for 5 weeks, the relative abundance of Alteromonadales increased relative to other orders with increasing AHL concentrations. On MBA alone, Alteromonadales was observed in the community of one replicate with a relative abundance of ca. 35%. In contrast, it was recovered from all three communities from MBA plates with 0.5 μM AHLs (MBAAHL0.5) where it dominated two replicates, and it was the dominant order on all three replicates from MBA plates with 50 μM AHLs (MBAAHL50). This trend was not observed for any of the other growth conditions. Most notable was the occurrence of Sphingobacteriales on substrates supplemented with AHLs. This order was not recovered from any of the other substrates, but one replicate plate in three of four treatments supplemented with 0.5 μM AHLs exhibited high relative abundances of this order (Fig. 6). The community structure analysis suggested that gelling agent and medium were stronger drivers of community composition than AHL supplementation, as the substrates containing AHLs seemed scattered among the other subclusters (Fig. 4). One exception was a distinct subcluster representing six MBA communities more similar to communities grown on SWA plates than to other MBA-derived communities. The parsimony test substantiated that differences in the community structures between AHL-supplemented and nonsupplemented substrates were not significant on the basis of the UniFrac distances, while the OTU-based analyses suggested that they were significant at an alpha level of 0.05 (Table 2). In situ concentrations of the four applied AHLs and 17 additional relevant AHLs (40) were below the detection level of 10 nM on both sampling occasions.
FIG 6.
Culturable community compositions on solid media supplemented with acylated homoserine lactones (AHLs) compared with the in situ composition and the compositions on nonsupplemented media. The compositions are depicted as relative proportions of bacterial orders obtained from seawater (5 μm, 0.2-μm filters) and from cultivated bacteria on marine broth agar (MBA) and marine broth agar supplemented with 0.5 (MBA•) or 50 (MBA••) μM AHLs (A), and seawater agar (SWA) and seawater agar supplemented with 0.5 (SWA•) or 50 (SWA••) μM AHLs (B).
DISCUSSION
Marine environments accommodate a wealth of different microbes, a diversity that is far from reflected in current collections of cultured marine bacteria. This lack of available laboratory cultures is a severe impediment to our understanding of the biology of marine microbes and also to the discovery of novel bioactive compounds. Based on previous findings (30–32, 34, 35, 37), we reasoned that replacing agar in different solid growth media targeting marine bacteria with an alternative gelling agent, i.e., gellan gum, would affect culturability. The findings presented in this study show that this substitution does improve culturability of marine bacteria, in terms of both viable cell counts and culturable diversity. In fact, we observed an increase in viable counts of 3- to 40-fold on substrates solidified with gellan gum compared with that from substrates solidified with agar. Cultivation of soil communities on gellan gum substrates has been shown to increase the viable count 1- to 2-fold (35), and similar results have been shown for lake sediment communities (37). The improvement observed in our study was most significant for the nutrient-poor seawater media, where viable counts were generally very low. Rich media supported the growth of more bacteria in all cases; most were observed on MBG substrates with 6.6% relative to direct counts. Hence, a rich medium solidified with gellan gum seems to be the most appropriate of the tested substrates to cultivate the majority of bacteria from these coastal surface waters. Incubation times of up to 3 months may improve the culturability of soil and marine bacteria (20, 21, 25, 41), yet in comparison with medium and gelling agent, incubation time and temperature had minor effects on viable counts in this study.
Data describing differences in the culturable diversity on substrates on the basis of different gelling agents are generally limited and based on very small data sets, yet previous studies have suggested that while gellan gum improves viable cell counts, the diversity of culturable soil bacteria may not be significantly different (25, 35, 38). In contrast, we found that the gelling agent was a key driver of the structure of the different cultured communities in both samplings. In fact, from S1, gelling agent was the only factor that had a significant effect on the diversity between cultured communities. This was evident from both the phylogenetic distances between samples (weighted UniFrac) and the community differences occurring at the species level (sequences binned at 97% sequence similarity). Furthermore, media solidified with gellan gum substrates recovered more of the OTUs that were observed in seawater than their corresponding agar-solidified substrates at both sampling time points. The fact that gelling agent indeed does affect culturable diversity corroborates observations by Tamaki et al. who saw a change in the relative abundance of phyla in randomly chosen colonies from agar- and gellan gum-based substrates inoculated with freshwater sediment samples (36).
Some of the major differences between the fractions of the communities that were culturable on gellan gum substrates and agar substrates related to the relative abundances of the orders of Flavobacteriales, Actinomycetales, Campylobacterales, Burkholderiales, Rhodospirillales, Rhodobacterales, and Thiotrichales. Flavobacteriales exhibited higher abundance on substrates solidified with agar; hence, conventional agar-based media may be more suited for targeting marine flavobacteria. The remaining orders were found in higher abundances, or exclusively, on media solidified with gellan gum, showing that gelling agent can be a powerful tool in the cultivation of targeted groups of marine bacteria. Moreover, sequences representing unknown bacterial orders were observed more frequently in communities derived from substrates solidified with gellan gum, showing that the cultured communities on gellan gum plates are not only different from communities culturable on agar-based substrates but also cover more of the natural community and more of the unknown fraction of this community. Some of the abundant unclassified OTUs were only related to environmental sequences, whereas others were closely related (>95% sequence similarity) to marine gammaproteobacteria and actinobacteria isolated using fluorescence-activated cell sorting (42). One of the most abundant OTUs representing an unknown order, OTU279, was a putative marine actinobacterium related to the novel subclass “Candidatus Actinomarinidae” (39). The facts that hitherto uncultured marine actinobacteria are culturable on MBG plates and that the actinomycetes present in the seawater at S1 seemed to be recovered on MBG are promising in terms of discovering novel bioactive compounds, as this group of bacteria is currently responsible for approximately 7,000 of the compounds in the dictionary of natural products (43) and for more than half the microbially derived antibiotics currently known (8). Gavrish et al. reported that collections of soil actinobacteria isolated using gellan gum were different from and more diverse than actinobacteria obtained using agar (44). Taken together, the results show that it will likely be fruitful for future bioprospecting efforts looking to obtain novel bioactive compounds from marine and soil actinobacteria to incorporate alternative gelling agents. As it is only a few families of the actinobacteria, e.g., the Streptomycetaceae and Micromonosporaceae, that produce the bulk of the bioactive compounds known from this group (8), it is uncertain whether gellan gum supports the growth of more bioactive strains. This will have to be assessed by a thorough evaluation of new isolates or by a metagenomics-based approach.
The reason for the altered culturability observed between gellan gum substrates and agar substrates is not known. However, agar and gellan gum are composed of different sugars, and agar, being a macroalgal polymer, may bias the composition of the cultured community toward certain microbes specialized in utilizing agar or, potentially, galactose residues released from the substrate. However, the similar observations from soil and freshwater environments (35–37) suggest that it may rather be the glucose, glucuronic acid, and rhamnose residues from gellan gum (45) that potentially stimulate the growth of a broader section of the microbial community. Alternatively, the agar media have negative effects on bacterial growth. In fact, it was recently shown that reactive oxygen species may be formed during sterilization of agar media, negatively affecting culturability (46).
We also assessed the effect of factors other than gelling agent on culturability. As mentioned, the medium had a significant impact on viable counts, whereas incubation time and temperature had less marked, yet in some cases measurable, effects on culturability. This was also true when considering community composition and the diversity of cultured communities. At the species level, temperature did not have a significant effect on community structure in any of the samplings, but the UniFrac data suggested that temperature may have had an effect during S2. We never observed a significant effect of incubation time on the clustering of individual culturable communities. However, it is well known that incubation times spanning months facilitate the cultivation of some fastidious bacteria (20, 21, 25, 41), and the lack of marked differences in our study is probably due to the relatively small temporal difference between treatments, i.e., a 3-week difference in incubation time at 25°C and 5 weeks at 10°C. From S2, we observed an interaction between gelling agent and medium. Specifically, all SWA-grown communities formed distinct subclusters. One reason why the communities from these plates were different from those grown on other media was that the Campylobacterales order, which dominated the seawater community and the other substrates, did not grow on these plates, highlighting that small-scale temporal variations in the natural community, the gelling agent, and the growth medium all significantly affect the outcome of cultivation efforts.
Moreover, we speculated that supplementing substrates with extracellular signaling molecules could enhance the culturability of marine bacteria. Previous attempts to culture Baltic Sea bacterioplankton in liquid cultures (47) suggested that micromolar concentrations of AHLs stimulate culturability. We supplemented media with AHLs known from marine Gram-negative bacteria, specifically of the Vibrionaceae family (40), but AHL supplementation had a negative impact on viable counts, suggesting that high concentrations of AHLs, compared with the in situ concentrations of <10 nM, may in fact be toxic to some bacteria. Furthermore, it did not have a clear effect on the culturable diversity. Efforts to culture marine sponge-associated bacteria on agar plates with AHLs have shown similar results (48); hence, increasing the culturability on solid media supplemented with signaling molecules is not straight forward. One exception was the Sphingobacteriales, which accounted for approximately 10% of sequences on some SWAAHL0.5 and MBAAHL0.5 plates. Thus, efforts to cultivate specific community members on solid substrates can be facilitated using AHLs.
In conclusion, it is clear that a substantial fraction of the hitherto unculturable marine bacteria are in fact culturable when the right substrates are applied. The gelling agent is specifically an important, but often overlooked, factor, and replacing agar with other gelling agents in future efforts to cultivate marine bacteria is essential for bringing more marine bacteria into culture and to discover new natural products.
MATERIALS AND METHODS
Sampling.
Surface seawater was collected at two different time points in Hornbæk Harbor, Denmark (56°05′42″N, 12°27′29″E), namely on 28 September 2015 at 8:30 a.m. (14.3°C, 4.85 mg liter−1 dissolved oxygen [DO], 14 practical salinity units [PSU]) for S1 and on 1 November 2015 at 9:30 a.m. (10.4°C, 0.81 mg liter−1 DO, 25 PSU) for S2. At each sampling, approximately 3 liters of seawater was collected at a depth of 0.5 m from three replicate sampling sites (A, B, and C) within a radius of 10 m.
In situ abundances of planktonic bacteria.
Total bacterial abundances in the free-living-size fraction (<5 μm) were determined using epifluorescence counts of SYBR gold-stained cells as previously described (49, 50).
Growth media and cultivation conditions.
Prefiltered seawater (5 μm) was plated on four different growth media (marine broth or seawater solidified with agar or gellan gum). Two of the media were also supplemented with two different concentrations of AHLs. Marine broth agar (MBA) and marine broth gellan gum (MBG) contained 37.4 g marine broth (BD Difco, Franklin Lakes, NJ, USA) supplemented with either 15 g agar (AppliChem, Darmstadt, Germany) or 6.5 g gellan gum (CP Kelco, San Diego, CA, USA) and 0.95 g MgSO4·7H2O per liter distilled H2O (dH2O). Seawater agar (SWA) and seawater gellan gum (SWG) contained either 15 g agar or 6.5 g gellan gum and 0.95 g MgSO4·7H2O per liter glass wool-filtered (GWF) seawater.
The ingredients for agar-containing media (MBA and SWA) were dissolved by boiling and the media were sterilized. SWG medium was prepared by mixing gellan gum, MgSO4·7H2O, and GWF seawater and then sterilized. The MBG medium was prepared in two solutions, one with 4× concentrated marine broth (14.96 g in 100 ml dH2O) and one with 2.6 g gellan gum and 0.39 g MgSO4·7H2O in 300 ml dH2O. The solutions were sterilized, cooled to 80°C, mixed, and immediately poured into plates. All growth media were sterilized by autoclaving at 121°C for 15 min.
The AHL-containing media (only MBA and SWA) were supplemented with two different concentrations of a mixture of four AHLs: N-(3-hydroxylhexanoyl)-l-homoserine lactone (HSL) (OH-C6; University of Nottingham, Nottingham, UK), N-(3-hydroxydecanoyl)-dl-HSL (OH-C10; Sigma, St. Louis, MO, USA), N-(3-oxodecanoyl)-l-HSL (O-C10; Sigma, St. Louis, MO, USA), and N-(3-oxododecanoyl)-l-HSL (O-C12; Sigma, St. Louis, MO, USA). Stock solutions with 10 mM of each AHL dissolved in ethyl acetate (EtOAc) were stored at −20°C. After sterilizing the media, equal concentrations (0.125 μM or 12.5 μM) of each of the AHLs were added, giving final concentrations of 0.5 μM or 50 μM, respectively. To minimize the inactivation of the compounds, the media were poured in petri dishes, dried, and immediately stored at 5°C. Furthermore, they were made within 24 h of use. All other media were supplemented with an amount of solvent (EtOAc) equivalent to that added to AHL-supplemented media.
The prefiltered seawater samples from replicate samples A, B, and C were 10-fold serially diluted in 0.9% saline solution and plated on each medium type. Four identical sets of plates (MBA, MBG, SWA, and SWG) were inoculated at each of the two samplings; one set of plates was incubated at 25°C for 2 weeks, one at 25°C for 5 weeks, one set at 10°C for 5 weeks, and one at 10°C for 10 weeks. To assess the effects of AHLs on culturability, four identical sets of 0.5 μM or 50 μM AHL-supplemented plates (MBAAHL0.5, MBAAHL50, SWAAHL0.5, and SWAAHL50) were also used for plating and incubated as above. The culturability (%) of marine bacteria on the respective media was based on CFU counts in comparison to the in situ bacterial abundances determined by fluorescence microscopy counts.
Detection of AHLs in seawater.
A 2-fold serial dilution of a mixture of the four AHLs (from 50 to 0.02 μM AHL mix) was prepared in filtered (GWF) autoclaved seawater. Each of the dilutions was extracted with equal volumes of EtOAc containing 0.5% formic acid (FA), shaken at 200 rpm for 30 min, and centrifuged at 3,000 × g for 5 min before the upper EtOAc phase was removed and evaporated to dryness under nitrogen flow. Samples were resuspended in 300 μl of acetonitrile (ACN)-H2O (1:1) with 1% (vol/vol) FA, vortexed, centrifuged for a short time, and transferred to autosampler vials. The samples (1 μl injected) were analyzed using ultra-high-performance liquid chromatography–quadrupole time of flight mass spectrometry (UHPLC-QTOF MS) on an Agilent 1290 Infinity UHPLC (Agilent Technologies, Santa Clara, CA) fitted with an Agilent Poroshell 120 phenyl-hexyl column (2.7 μm, 250 mm by 2.1 mm) and coupled to an Agilent 6545 QTOF, as described in detail in reference 51. A linear gradient consisting of water and ACN both buffered with 20 mM FA and the water further buffered with 10 mM ammonium formate was used. The gradient started at 5% ACN and increased to 85% in 14 min, and then to 100% in 1 min where it was held for 2 min. Data processing was performed using MassHunter Qualitative Analysis B.07.00 (Agilent Technologies) using a mass extraction window of m/z ± 0.005 for the [M + H]+, [M + Na]+, and [M + NH4]+ ions of the AHLs.
Besides the four AHLs used in the media, the following reference standards were coanalyzed in the sequence: N-butanoyl-HSL (C4), N-hexanoyl-HSL (C6), N-octanoyl-HSL (C8), N-decanoyl-HSL (C10), N-dodecanoyl-HSL (C12), N-tetradecanoyl-HSL (C14), N-(3-oxohexanoyl)-HSL (O-C6), and N-(3-oxooctanoyl)-HSL (O-C8).
DNA extraction and V4 amplicon sequencing.
To compare the microbial community in seawater with the culturable microbial communities on different solid media, DNA was extracted for V4 amplicon sequencing. At each sampling time point, particle-associated bacteria from 3 liters of seawater were filtered through two 5-μm polycarbonate (PC) filters (8-μm mixed cellulose ester [MCE] backing filter; GE Water & Process Tech., Trevose, PA, USA) (1.5 liter per filter). The planktonic fractions in the filtrates were subsequently filtered through three 0.2-μm PC filters (Frisenette, Knebel, Denmark) with 1-μm MCE backing filters (Advantec, Dublin, CA, USA), 1 liter on each filter. All filters were stored at −20°C in 50-ml Falcon tubes until DNA was extracted. Prior to extraction, each filter was divided into 5-mm strips, and 5 ml lysis buffer (40 mM EDTA, 50 mM Tris [pH 8.3], and 0.75 M sucrose) was added to each tube. Bacterial biomass from solid substrate plates was harvested from three replicate plates containing approximately 200 separable macrocolonies each. Biomass was harvested by washing the plates with lysis buffer (5 ml per plate) and transferring the solutions to 50-ml Falcon tubes.
Filters and colony biomass from plates were incubated at room temperature for 1 h with regular agitation. Samples were heated to 100°C for 10 min and cooled. The rest of the DNA extraction procedure was adapted from that described in reference 52. Lysozyme (Sigma, St. Louis, MO, USA) was added to a final concentration of 100 mg ml−1 and samples were incubated at 37°C for 45 min with regular agitation. Sodium dodecyl sulfate (SDS) and proteinase K (Sigma, St. Louis, MO, USA) were added to final concentrations of 0.4% and 0.2 mg ml−1, respectively. After incubating for 1 h at 55°C with regular agitation, samples were centrifuged at 3,000 × g for 5 min and transferred to new tubes. Extractions were performed twice with 5 ml of phenol-chloroform-isoamyl alcohol (25:24:1 vol/vol/vol; Sigma, St. Louis, MO, USA), followed by an extraction of the aqueous phase with one volume of chloroform-isoamyl alcohol (24:1 vol/vol; Sigma, St. Louis, MO, USA). Ethanol precipitation was performed using 2 volumes of ice-cold 96% ethanol and 0.1 volume of sodium acetate (3 M, pH 5.5) and incubating at −20°C overnight. The precipitated DNA was pelleted, washed with ethanol, and dissolved in sterile Milli-Q water (pH 8).
A total of 106 samples were included in the V4 amplicon sequencing, 29 samples from S1 representing five seawater samples and 24 plate communities and 77 samples from S2 representing five seawater samples and 72 plate communities. A nested PCR approach was applied (53, 54), in which the 16S rRNA genes were amplified using the universal 27F and 1492R primer set (55). The V4 region was amplified using the 16S rRNA gene PCR products as the template in the subsequent PCR applying primers listed in Table 3 and procedures previously described (53). The PCR products were purified (AmPure XP PCR purification; Agencourt Bioscience Corporation, Beverly, MA, USA), quantified (Qubit dsDNA BR assay; Eugene, USA), and pooled (3 μg PCR product from each sample) prior to 250PE Illumina MiSeq sequencing at BGI Tech Solutions, Hong Kong.
TABLE 3.
V4 primers for 250PE Illumina MiSeq sequencing used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| Forward | |
| V4.SA501 | AATGATACGGCGACCACCGAGATCTACACATCGTACGTATGGTAATTGTGTGCCAGCMGCCGCGGTAA |
| V4.SA502 | AATGATACGGCGACCACCGAGATCTACACACTATCTGTATGGTAATTGTGTGCCAGCMGCCGCGGTAA |
| V4.SA503 | AATGATACGGCGACCACCGAGATCTACACTAGCGAGTTATGGTAATTGTGTGCCAGCMGCCGCGGTAA |
| V4.SA504 | AATGATACGGCGACCACCGAGATCTACACCTGCGTGTTATGGTAATTGTGTGCCAGCMGCCGCGGTAA |
| V4.SA505 | AATGATACGGCGACCACCGAGATCTACACTCATCGAGTATGGTAATTGTGTGCCAGCMGCCGCGGTAA |
| V4.SA506 | AATGATACGGCGACCACCGAGATCTACACCGTGAGTGTATGGTAATTGTGTGCCAGCMGCCGCGGTAA |
| V4.SA507 | AATGATACGGCGACCACCGAGATCTACACGGATATCTTATGGTAATTGTGTGCCAGCMGCCGCGGTAA |
| V4.SA508 | AATGATACGGCGACCACCGAGATCTACACGACACCGTTATGGTAATTGTGTGCCAGCMGCCGCGGTAA |
| Reverse | |
| V4.SA701 | CAAGCAGAAGACGGCATACGAGATAACTCTCGAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT |
| V4.SA702 | CAAGCAGAAGACGGCATACGAGATACTATGTCAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT |
| V4.SA703 | CAAGCAGAAGACGGCATACGAGATAGTAGCGTAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT |
| V4.SA704 | CAAGCAGAAGACGGCATACGAGATCAGTGAGTAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT |
| V4.SA705 | CAAGCAGAAGACGGCATACGAGATCGTACTCAAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT |
| V4.SA706 | CAAGCAGAAGACGGCATACGAGATCTACGCAGAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT |
Index sequences are shown in bold.
Sequence analyses.
Demultiplexed sequences were assembled and denoised using mothur v.1.33.3 (56). Poorly assembled sequences, sequences containing ambiguous base calls, sequences containing homopolymers longer than eight nucleotides, and sequences aligning poorly to the SILVA database (version 123) (51) were excluded from the data sets. Chimeras were identified using UCHIME (57), and these were removed along with sequences of nonbacterial origin. OTU-based and phylogeny-based approaches were applied in the sequence analysis. In OTU-based analysis, the clean sequences were clustered into operational taxonomic units (OTUs) using a species-level cutoff (97% nucleotide sequence similarity) after which the OTUs were classified according to the SILVA database. Reproducibility between replicate samples and recovery of seawater OTUs from the different media was determined based on 8,500 sequences per sample (rarefaction curves are in Fig. S1 in the supplemental material) and visualized using R v. 3.3.2 and the Vennerable package. Sequences that were unclassified at the order level and were highly abundant on solid media were subjected to a nucleotide BLAST (BLASTn) against the NCBI nucleotide sequence database. Community composition analyses were performed using the phyloseq package in R v. 3.3.2. Measures of alpha (Chao1 and Shannon) and beta (Bray-Curtis distances) diversity were also calculated based on 8,500 sequences from each sample. In the phylogeny-based approach, beta-diversity was assessed using weighted UniFrac distances calculated using the weighted UniFrac algorithm from mothur v.1.33.3. Heatmaps showing relative OTU richness were constructed using the CIMminer tool for clustered image maps (https://discover.nci.nih.gov/cimminer/).
Statistical analyses.
To evaluate the significance of the observed differences in overall culturability on growth media solidified with agar and gellan gum, paired two-tailed t tests were applied (n = 8, α = 0.05).
The significance of the differences in microbial community structure (beta-diversity) were assessed using AMOVA (58–60) on Bray-Curtis distances from the OTU-based sequence analysis (1,000 randomizations, α = 0.05) and using the parsimony method (P test) on the weighted UniFrac distances in the phylogeny-based analysis (1,000 randomizations, α = 0.05).
Accession number(s).
Amplicon sequencing reads were deposited in the NCBI Sequence Read Archive (SRA) database (accession number SRP091481) and a representative sequence of the unclassified actinobacterial OTU279 was deposited in GenBank under accession number KY452456.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the MaCuMBA Project under the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 311975 and by grant VKR023285 from the Villum Foundation.
Agilent Technologies is acknowledged for providing UHPLC-QTOF MS from the Thought Leader Award.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00243-17.
REFERENCES
- 1.Joint I, Mühling M, Querellou J. 2010. Culturing marine bacteria—an essential prerequisite for biodiscovery. Microb Biotechnol 3:564–575. doi: 10.1111/j.1751-7915.2010.00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bull AT, Ward AC, Goodfellow M. 2000. Search and discovery strategies for biotechnology: the paradigm shift. Microbiol Mol Biol Rev 64:573–606. doi: 10.1128/MMBR.64.3.573-606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glöckner FO, Joint I. 2010. Marine microbial genomics in Europe: current status and perspectives. Microb Biotechnol 3:523–530. doi: 10.1111/j.1751-7915.2010.00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fenical W. 1993. Chemical studies of marine bacteria: developing a new resource. Chem Rev 93:1673–1683. doi: 10.1021/cr00021a001. [DOI] [Google Scholar]
- 5.Engel S, Jensen PR, Fenical W. 2002. Chemical ecology of marine microbial defense. J Chem Ecol 28:1971–1985. doi: 10.1023/A:1020793726898. [DOI] [PubMed] [Google Scholar]
- 6.Lewenza S, Visser MB, Sokol PA. 2002. Interspecies communication between Burkholderia cepacia and Pseudomonas aeruginosa. Can J Microbiol 48:707–716. doi: 10.1139/w02-068. [DOI] [PubMed] [Google Scholar]
- 7.Lovell FM. 1966. The structure of a bromine-rich marine antibiotic. J Am Chem Soc 88:4510–4511. doi: 10.1021/ja00971a040. [DOI] [Google Scholar]
- 8.Murphy BT, Jensen PR, Fenical W. 2012. The chemistry of marine bacteria, p 153–190. In Fattorusso E, Gerwick W, Taglialatela-Scafati O (ed), Handbook of marine natural products. Springer Science+Business Media B.V., Dordrecht, Netherlands. [Google Scholar]
- 9.Staley JT, Konopka A. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T, Somerfield PJ, Huse S, Joint I. 2009. The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- 11.Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 12.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y-H, Smith HO. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 13.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM. 2005. Comparative metagenomics of microbial communities. Science 308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 14.DeLong EF. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 15.Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S, Binder BJ, Edmonds J, Ye W, Orcutt B, Howard EC, Meile C, Palefsky W, Goesmann A, Ren Q, Paulsen I, Ulrich LE, Thompson LS, Saunders E, Buchan A. 2007. Ecological genomics of marine roseobacters. Appl Environ Microbiol 73:4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, McDaniel L, Moran MA, Nelson KE, Nilsson C, Olson R, Paul J, Brito BR, Ruan Y, Swan BK, Stevens R, Valentine DL, Thurber RV, Wegley L, White BA, Rohwer F. 2008. Functional metagenomic profiling of nine biomes. Nature 452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P, Joint I. 2008. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steele HL, Jaeger K-E, Daniel R, Streit WR. 2009. Advances in recovery of novel biocatalysts from metagenomes. J Mol Microbiol Biotechnol 16:25–37. doi: 10.1159/000142892. [DOI] [PubMed] [Google Scholar]
- 19.Mühling M, Joint I, Willetts AJ. 2013. The biodiscovery potential of marine bacteria: an investigation of phylogeny and function. Microb Biotechnol 6:361–370. doi: 10.1111/1751-7915.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song J, Oh H-M, Cho J-C. 2009. Improved culturability of SAR11 strains in dilution-to-extinction culturing from the East Sea, West Pacific Ocean. FEMS Microbiol Lett 295:141–147. doi: 10.1111/j.1574-6968.2009.01623.x. [DOI] [PubMed] [Google Scholar]
- 21.Bentzon-Tilia M, Farnelid H, Jürgens K, Riemann L. 2014. Cultivation and isolation of N2-fixing bacteria from suboxic waters in the Baltic Sea. FEMS Microbiol Ecol 88:358–371. doi: 10.1111/1574-6941.12304. [DOI] [PubMed] [Google Scholar]
- 22.Farnelid H, Harder J, Bentzon-Tilia M, Riemann L. 2014. Isolation of heterotrophic diazotrophic bacteria from estuarine surface waters. Environ Microbiol 16:3072–3082. doi: 10.1111/1462-2920.12335. [DOI] [PubMed] [Google Scholar]
- 23.Tyson GW, Lo I, Baker BJ, Allen EE, Hugenholtz P, Banfield JF. 2005. Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp. nov. from an acidophilic microbial community. Appl Environ Microbiol 71:6319–6324. doi: 10.1128/AEM.71.10.6319-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nichols D, Lewis K, Orjala J, Mo S, Ortenberg R, O'Connor P, Zhao C, Vouros P, Kaeberlein T, Epstein SS. 2008. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl Environ Microbiol 74:4889–4897. doi: 10.1128/AEM.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis KER, Joseph SJ, Janssen PH. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microbiol 71:826–834. doi: 10.1128/AEM.71.2.826-834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Epstein SS. 2013. The phenomenon of microbial uncultivability. Curr Opin Microbiol 16:636–642. doi: 10.1016/j.mib.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 27.D'Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K. 2010. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol 17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang KS, Veeder GT, Mirrasoul PJ, Kaneko T, Cottrell IW. 1982. Agar-like polysaccharide produced by a pseudomonas species: production and basic properties. Appl Environ Microbiol 43:1086–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris ER, Nishinari K, Rinaudo M. 2012. Gelation of gellan—a review. Food Hydrocoll 28:373–411. doi: 10.1016/j.foodhyd.2012.01.004. [DOI] [Google Scholar]
- 30.Shungu D, Valiant M, Tutlane V, Weinberg E, Weissberger B, Koupal L, Gadebusch H, Stapley E. 1983. GELRITE as an agar substitute in bacteriological media. Appl Environ Microbiol 46:840–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin CC, Casida LE Jr. 1984. GELRITE as a gelling agent in media for the growth of thermophilic microorganisms. Appl Environ Microbiol 47:427–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris JE. 1985. GELRITE as an agar substitute for the cultivation of mesophilic Methanobacterium and Methanobrevibacter species. Appl Environ Microbiol 50:1107–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marteinsson VT, Birrien J-L, Prieur D. 1997. In situ enrichment and isolation of thermophillic microorganisms from deep-sea vent environments. Can J Microbiol 43:694–697. doi: 10.1139/m97-100. [DOI] [Google Scholar]
- 34.Rule PL, Alexander AD. 1986. Gellan gum as a substitute for agar in leptospiral media. J Clin Microbiol 23:500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol 68:2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamaki H, Sekiguchi Y, Hanada S, Nakamura K, Nomura N, Matsumura M, Kamagata Y. 2005. Comparative analysis of bacterial diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation-based techniques. Appl Environ Microbiol 71:2162–2169. doi: 10.1128/AEM.71.4.2162-2169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamaki H, Hanada S, Sekiguchi Y, Tanaka Y, Kamagata Y. 2009. Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environ Microbiol 11:1827–1834. doi: 10.1111/j.1462-2920.2009.01907.x. [DOI] [PubMed] [Google Scholar]
- 38.Sait M, Hugenholtz P, Janssen PH. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ Microbiol 4:654–666. doi: 10.1046/j.1462-2920.2002.00352.x. [DOI] [PubMed] [Google Scholar]
- 39.Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F. 2013. Metagenomics uncovers a new group of low GC and ultra-small marine Actinobacteria. Sci Rep 3:2471. doi: 10.1038/srep02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen B, Nielsen K, Machado H, Melchiorsen J, Gram L, Sonnenschein E. 2014. Global and phylogenetic distribution of quorum sensing signals, acyl homoserine lactones, in the family of Vibrionaceae. Mar Drugs 12:5527–5546. doi: 10.3390/md12115527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connon SA, Giovannoni SJ. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol 68:3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Garcia M, Brazel DM, Swan BK, Arnosti C, Chain PSG, Reitenga KG, Xie G, Poulton NJ, Gomez ML, Masland DED, Thompson B, Bellows WK, Ziervogel K, Lo C-C, Ahmed S, Gleasner CD, Detter CJ, Stepanauskas R. 2012. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS One 7:e35314. doi: 10.1371/journal.pone.0035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen PR, Mincer TJ, Williams PG, Fenical W. 2005. Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek 87:43–48. doi: 10.1007/s10482-004-6540-1. [DOI] [PubMed] [Google Scholar]
- 44.Gavrish E, Bollmann A, Epstein S, Lewis K. 2008. A trap for in situ cultivation of filamentous actinobacteria. J Microbiol Methods 72:257–262. doi: 10.1016/j.mimet.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollock TJ. 1993. Gellan-related polysaccharides and the genus Sphingomonas. J Gen Microbiol 139:1939–1945. doi: 10.1099/00221287-139-8-1939. [DOI] [Google Scholar]
- 46.Tanaka T, Kawasaki K, Daimon S, Kitagawa W, Yamamoto K, Tamaki H, Tanaka M, Nakatsu CH, Kamagata Y. 2014. A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl Environ Microbiol 80:7659–7666. doi: 10.1128/AEM.02741-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruns A, Cypionka H, Overmann J. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl Environ Microbiol 68:3978–3987. doi: 10.1128/AEM.68.8.3978-3987.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sipkema D, Schippers K, Maalcke WJ, Yang Y, Salim S, Blanch HW. 2011. Multiple approaches to enhance the cultivability of bacteria associated with the marine sponge Haliclona (gellius) sp. Appl Environ Microbiol 77:2130–2140. doi: 10.1128/AEM.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noble R, Fuhrman J. 1998. Use of SYBR green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol 14:113–118. doi: 10.3354/ame014113. [DOI] [Google Scholar]
- 50.Gram L, Melchiorsen J, Bruhn JB. 2010. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar Biotechnol (NY) 12:439–451. doi: 10.1007/s10126-009-9233-y. [DOI] [PubMed] [Google Scholar]
- 51.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boström KH, Simu K, Hagström Å, Riemann L. 2004. Optimization of DNA extraction for quantitative marine bacterioplankton community analysis. Limnol Oceanogr Methods 2:365–373. doi: 10.4319/lom.2004.2.365. [DOI] [Google Scholar]
- 53.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ. 2015. Evaluation of water sampling methodologies for amplicon-based characterization of bacterial community structure. J Microbiol Methods 114:43–50. doi: 10.1016/j.mimet.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 55.Lane D. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. J. Wiley and Sons, Amsterdam, Netherlands. [Google Scholar]
- 56.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 60.Martin AP. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol 68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.