FIG 1.
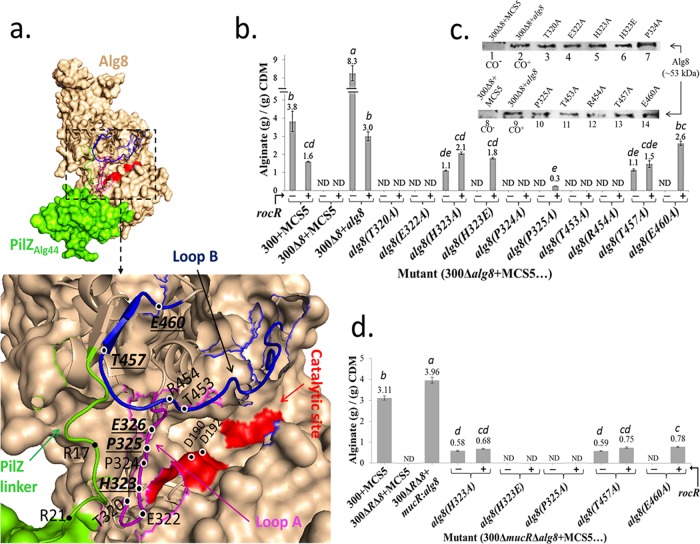
Highly conserved amino acids of Alg8 are involved in c-di-GMP-dependent regulation of alginate polymerization. (a) The in silico fusion of Alg8-PilZAlg44 was modeled using the Phyre2 server. Residues selected for site-specific mutagenesis were shown on loop A (magenta), loop B (blue), and the PilZ domain (green). Mutations of bolded and underlined residues were responsive to the absence and presence of RocR overproduction (i.e., reduced levels of c-di-GMP), while for other shown residues alginate production was abolished independent of RocR. (b) Alginate quantification of PDO300Δalg8 transformants harboring various plasmids containing respective site-specific mutants of alg8 with (+) and without (−) the rocR gene. (c) Immunoblot analysis of envelope fractions developed using an anti-Alg8 antibody showed that none of mutations affected the Alg8 localization to the envelope fraction (lanes 3 to 7 and lanes 10 to 14). Lanes 1 to 8 and 2 to 9 represent negative and positive controls, respectively. For estimating relative protein amounts, the protein band intensity was analyzed by the ImageJ software. The protein band based on genomic expression derived from 300Δ8+alg8 (lanes 2 and 9) was set as 1.0 in density, and relative densities of other bands were calculated as follows: lane 3 (1.14), lane 4 (0.937), lane 5 (1.06), lane 6 (1.03), lane 7 (1.48), lane 10 (0.9), lane 11 (0.848), lane 12 (0.817), lane 13 (0.985), and lane 14 (0.987). (d) Highly conserved amino acids of Alg8 whose replacement with alanine decoupled alginate polymerization from c-di-GMP-dependent and MucR-dependent regulation. Alginate quantification was performed for PDO300ΔmucRΔalg8 transformants with plasmids harboring respective site-specific mutants of alg8 with (+) and without (−) the rocR gene. The data in histograms in panels b and d represent the means ± SD for four independent repetitions, and treatments with different lowercase italic letters above the bars are significantly different (post hoc Tukey's HSD test, P < 0.05). CDM, cell dry mass; 300, PDO300; ND, not detectable; MCS5, pBBR1MCS-5.