Figure 4. Percentage ancestry based on ancestry informative markers (AIMs) in Anopheles gambiae individuals from Guinea Bissau.
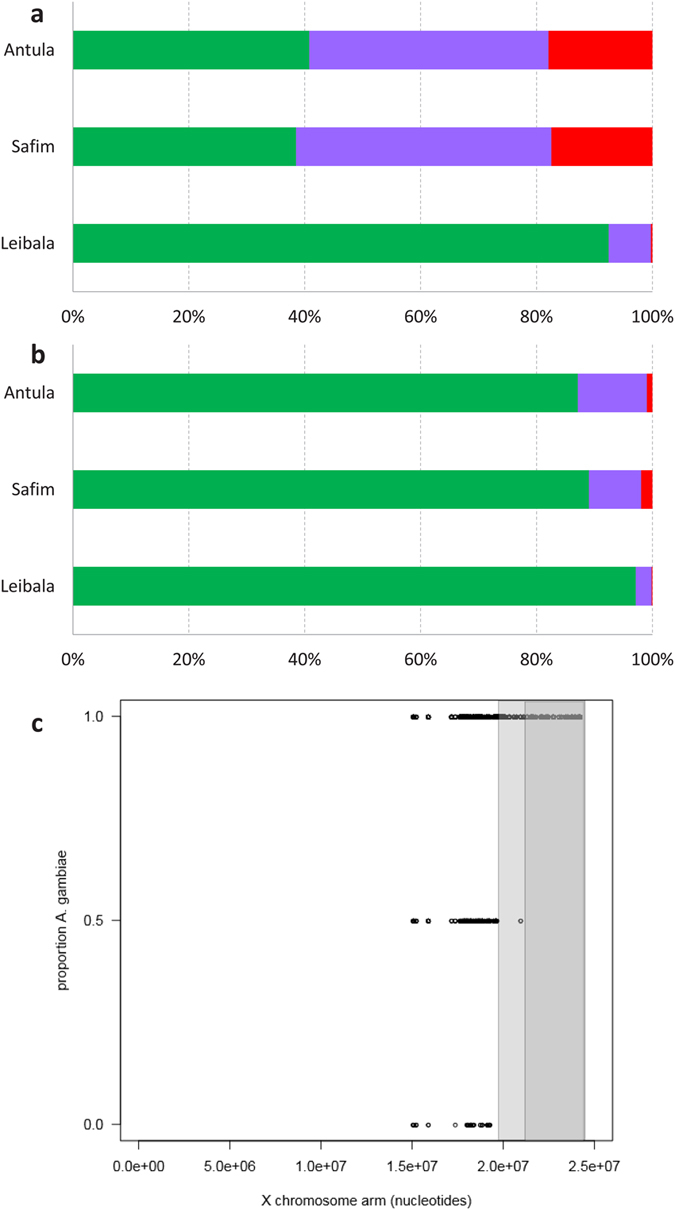
Stacked bars show percentage of ancestry found in whole genome sequenced individuals sampled in two coastal (Safim N = 5, Antula N = 12) and one inland locality (Leibala, N = 4) estimated using AIMs identified from genome-wide comparisons between samples from a more typical region of infrequent hybridization17. Percentage ancestry is based on 93 autosomal (a) and 236 chromosome-X (b) AIMs, each scored as being homozygous for A. gambiae (green) or A. coluzzii (red) or heterozygous/admixed (purple). (c) Position and genotype of chromosome-X AIMs. Each AIM was genotyped as 0 = A. coluzzii homozygote, 0.5 = A. coluzzii/gambiae heterozygote or 1 = A. gambiae homozygote. Shaded areas correspond to the >3 Mb region (dark grey) homozygous for for A. gambiae AIMs, which increases to >4 Mb (light grey) if a single heterozygous AIM is included.
