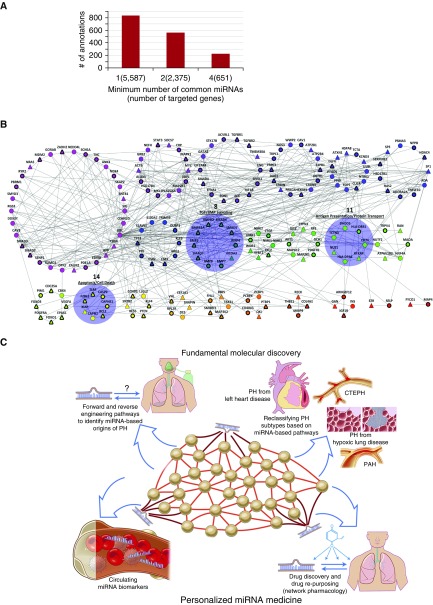
Figure 3.
Application of network theory to gain insight into microRNA (miRNA) activity in pulmonary arterial hypertension (PAH). (A) Gene set enrichment analysis (GSEA) demonstrates the vast pleiotropy of miRNA activity. The number of significant pathway annotations was gathered after GSEA enrichment on genes targeted by at least 1, 2, or 4 pulmonary hypertension (PH)-relevant miRNAs (among a group of 25 total miRNAs). A large number of statistically significant annotations was observed in all cases, indicating the need for additional statistical filtering techniques to discern the convergent functions of miRNAs. (B) Network architecture analysis improves the resolution of testable hypotheses regarding convergent miRNA function. From genes targeted by four or more PH-relevant miRNAs and overlaid on an extended network of PH genes and their first-degree interactors, a single connected component of 190 genes was partitioned into clusters based on its topology, using the “MAP” algorithm (58). Each node represents a gene, and each gray line denotes an interaction between genes. Rings of same-colored nodes represent clusters. Triangular nodes are those genes targeted by four or more PH-relevant miRNAs, and circular nodes are first-degree interactors necessary to make a single connected component. Nodes outlined in thick black lines are genes already known to play a role in PH, curated from the scientific literature. Of the 22 clusters, we found 3 to be of particular interest (highlighted in blue). Clusters 8, 11, and 14 are involved in transforming growth factor/bone morphogenetic protein (TGF/BMP) signaling, antigen presentation and protein transport, and apoptosis and cell death, respectively. (C) Leveraging computational gene network analysis for discovery of fundamental miRNA biology in PH and application to personalized medicine initiatives in PH. CTEPH = chronic thromboembolic pulmonary hypertension.