Figure 1.
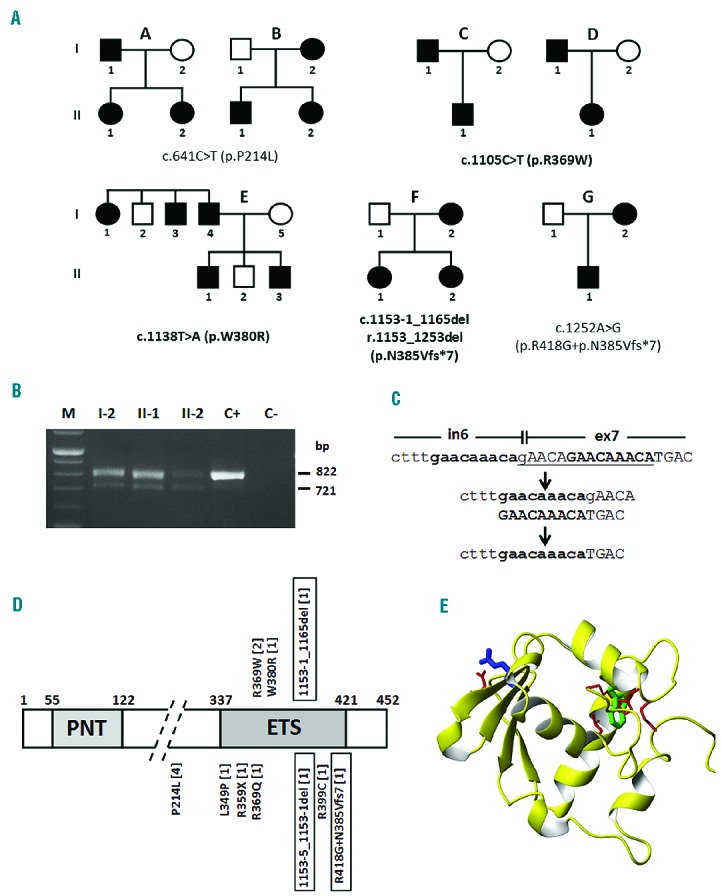
Mutations identified in the ETV6 gene and their effect on protein structure. (A) Pedigrees of families enrolled in this study carrying different mutations as indicated (novel mutations in bold). Nucleotide numbering reflects the ETV6 cDNA with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence (RefSeq NM_001987.4). Therefore, the initiation codon is residue 1 in the amino acid sequence. Families B and G have been previously reported (Noetzli et al.4). (B) RT-PCR in affected members (I-2, II-1, and II-2) of family F to determine the consequence of the c.1153-1_1165del mutation on splicing. C+, wild-type control; C−, negative control. The analysis shows two fragments, the wild-type (822 bp) and the exon 7 skipping (721 bp) products. (C) The deletion of the 14 bp (gAACAGAACAAACA) of c.1153-1_1165del is likely due to non-allelic homologous recombination between the two GAACAAACA repeats located at the intron 6 and exon 7 boundary. (D) Domain structure of ETV6 (XP_011518909.1) based on Pfam annotation at http://www.ncbi.nlm.nih.gov/gene/2120 (PNT, pointed N-terminal domain; ETS, C-terminal DNA binding domain), with mutations identified in ETV6, already reported or identified in this study (top). The numbers of families carrying each mutation are in brackets. Mutations leading to skipping of exon 7 are boxed. (E) Structural modeling of the ETS domain with residues R369 (blue) and W380 (green) affected by the p.R369W and p.W380R mutations.
