Figure 2.
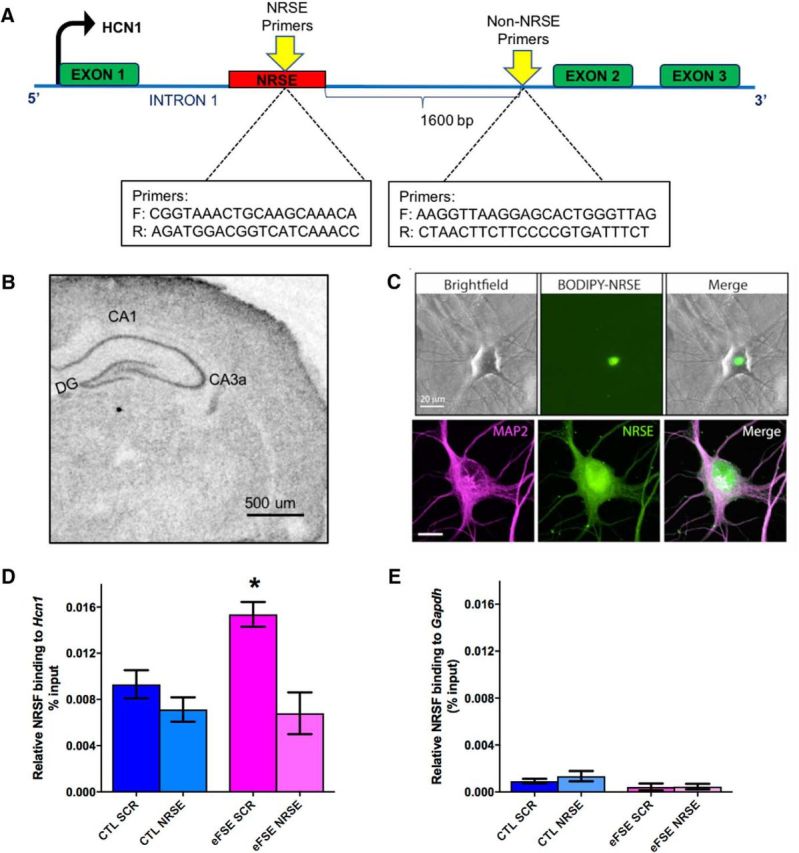
Augmented binding of NRSF to a target marker gene is specific, selective, increased after eFSE, and abrogated by a decoy strategy. A, Schematic showing the gene-structure and binding sites for NRSF (NRSF recognition element [NRSE]) within a target-gene, hcn1, and of a control region lacking an NRSE. B, In situ hybridization performed on a P11 rat brain section, demonstrating endogenous expression of NRSF in the hippocampus. A classical neuronal distribution is apparent. Scale bar, 500 μm. C, To interfere with NRSF chromatin binding after eFSE, we used a decoy strategy using ODNs with a sequence of the NRSE of hcn1. The ODNs enter hippocampal neurons grown in culture and their nuclei, as apparent by the use of ODNs tagged with the green fluorescent dye BODIPY. Top, Unstained neurons shown in bright-field, under green fluorescence setting (for visualizing the BODIPY) and as a combined image. Bottom, Neurons were subjected to immunohistochemistry for the dendritic marker MAP2, then imaged with both red and green fluorescence filters (left and middle), as well as combined. D, The use of ChIP demonstrated that NRSF occupancy at the hcn1 gene was not different between control rats that were given a scrambled ODN (CTL-SCR) and controls administered NRSE-ODN (CTL-NRSE; t(20) = 1.04; p > 0.05; Bonferroni's multiple-comparisons test). However, NRSF binding to HCN1 was significantly augmented in hippocampi of eFSE-SCR rats 3 h after eFSE (t(20) = 3.12, p < 0.05). Administration of NRSE-ODN after eFSE significantly reduced NRSF occupancy at the gene hcn1 in the eFSE-NRSE group compared with eFSE-SCR (t(20) = 4.07, p < 0.05), and this binding was not significantly different from controls. E, The specificity of NRSF binding to the NRSE is apparent from the minimal binding of the repressor to a gene (Gapdh) that lacks an NRSE. *p < 0.05.
