Figure 4.
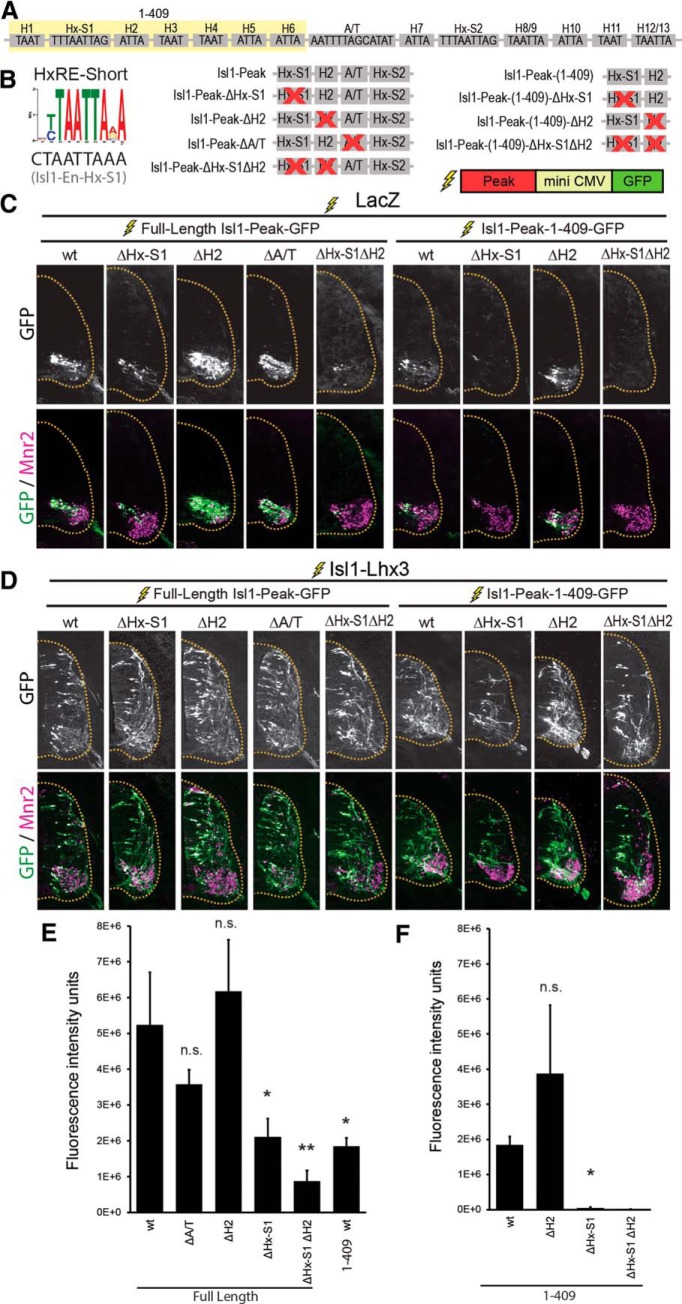
The Isl1-Peak is activated by the Isl1-Lhx3 complex. A, Schematic representation of HxRE-S1, HxRE-S2, 13 TAAT motifs, and A/T-rich motif within the Isl1-Peak. Yellow shading indicates the sequences included in the shortened Isl1-Peak-(1-409). B, HxRE-Short sequence identified by ChIP-Seq de novo motif analysis, and the HxRE-S1 sequences in Isl1-Peak. Isl1-Peak mutants used for GFP-reporter experiments. C, GFP-reporter experiments for Isl1-Peak variants. Embryos were electroporated with Isl1-Peak-GFP reporter constructs plus ubiquitously expressing LacZ to mark electroporated cells. Sections were immunostained for Mnr2 to mark MNs. Images are representative of electroporations from multiple embryos. Isl1-Peak-wt: n = 5, Isl1-Peak-ΔHx-S1: n = 6, Isl1-Peak-ΔH2: n = 5, Isl1-Peak-ΔA/T: n = 16, Isl1-Peak-ΔHx-S1ΔH2: n = 6. Isl1-Peak-(1-409)-wt: n = 17, Isl1-Peak-(1-409)-ΔHx-S1: n = 10, Isl1-Peak-(1-409)-ΔH2: n = 9, Isl1-Peak-(1-409)-ΔHx-S1ΔH2: n = 20. D, Embryos electroporated with an Isl1-Peak-GFP reporter construct plus Isl1-Lhx3 fusion protein construct. Sections immunostained for GFP and Mnr2. Images are representative of electroporations from multiple embryos. Isl1-Peak-wt: n = 6, Isl1-Peak-ΔHx-S1: n = 2, Isl1-Peak-ΔH2: n = 5, Isl1-Peak-ΔA/T: n = 3, Isl1-Peak-ΔHx-S1ΔH2: n = 4 . Isl1-Peak-(1-409)-wt: n = 5, Isl1-Peak-(1-409)-ΔHx-S1: n = 4, Isl1-Peak-(1-409)-ΔH2: n = 6, Isl1-Peak-(1-409)-ΔHx-S1ΔH2: n = 6. E, F, GFP fluorescence intensity for embryos electroporated with Isl1-Peak-GFP reporter constructs + LacZ; n = 4-12 embryos per condition. Results were analyzed with a one-way ANOVA followed by Holm multiple comparison analysis, comparing each mutant reporter construct to full-length wt-Isl1-Peak-GFP reporter or (E) wt-(1-409)-Isl1-Peak (F), *p < 0.05, **p < 0.01. Error bars represent the SEM.