Abstract
Importance
Whites have a higher risk of atrial fibrillation (AF) relative to Blacks, despite a lower prevalence of risk factors. This difference may be due, at least in part, to genetic factors.
Objective
To determine whether 9 single nucleotide polymorphisms (SNPs) associated with AF account for this paradoxical differential racial risk. We also used admixture mapping to search genome wide for loci that may account for this phenomenon.
Design, Setting, and Participants
Genome wide admixture analysis and candidate SNP study involving 3 population-based cohort studies initiated between 1987 and 1997, including the Cardiovascular Health Study (CHS; n=3,969), the Atherosclerosis Risk in Communities Study (ARIC; n=12,341), and the Health, Aging, and Body Composition Study (Health ABC; n=1,015).
Main Outcomes and Measures
Incident AF systematically ascertained using clinic visit ECGs, hospital discharge diagnosis codes, death certificates and Medicare claims data.
Results
Cox proportional hazards models and the proportion of treatment effect method were utilized to determine the impact of 9 AF-risk SNPs among participants from CHS and ARIC. A single SNP, rs10824026 (chromosome 10: position 73661450), was found to significantly mediate 11.4% (95% CI 2.9–29.9%) and 31.7% (95% CI 16.0–53.0%) of the higher risk in Whites compared to Blacks in CHS and ARIC, respectively. Admixture mapping was performed in a meta-analysis of Black participants within CHS (n=811), ARIC (n=3,112), and Health ABC (n=1,015). No loci that reached the pre-specified statistical threshold for genome-wide significance were identified.
Conclusions and Relevance
The rs10824026 SNP on chromosome 10q22 mediates a modest proportion of the increased risk of AF among Whites relative to Blacks, potentially through an impact on gene expression levels of MYOZ1. No additional genetic variants accounting for a significant portion of the differential racial risk of AF were identified with genome wide admixture mapping, suggesting that additional genetic or environmental influences beyond single SNPs in isolation may account for the paradoxical racial risk of AF among Whites and Blacks.
Journal Subject Codes: atrial fibrillation, race, genetics, admixture
Introduction
Atrial fibrillation (AF), the most common sustained cardiac arrhythmia, is a rapidly growing health epidemic associated with increased risks of heart failure, stroke, and death.1–3 Whites have an increased risk of AF relative to Blacks, despite a lower prevalence of well-established clinical risk factors.4–7 The paradoxical differential racial risk of AF among Whites and Blacks may be secondary to genetic factors. American Blacks are an admixed population with both African and European ancestry. We have shown that higher European ancestry among Blacks was associated with an increased risk of AF.8 This finding supports the possibility of genetic factors contributing to the increased risk of AF among Whites; however, the potential genetic culprits accounting for this association remain unknown.
To explore the genetics underlying the differential racial risk of the arrhythmia, we evaluated the impact of 9 SNPs previously associated with AF risk.9–14 We also sought to identify novel genetic loci associated with the arrhythmia through the use of admixture mapping. Admixture mapping is a modified form of a genome wide association study (GWAS) that can be utilized to uncover genetic loci in populations of mixed ancestry when there is a differential ancestral risk of disease (a more detailed description of admixture mapping is provided in the Online Supplement, eMethod1), and has been used to map loci and genetic variants for end-stage renal disease among African Americans, prostate cancer in African Americans, ethnic neutropenia in African Americans, breast cancer risk in Latinas, and other traits.15–21 Thus, we used genome wide admixture mapping in the African American populations in the Cardiovascular Health Study (CHS), the Atherosclerosis Risk in Communities Study (ARIC), and Health, Aging, and Body Composition Study (Health ABC) cohorts to search for novel loci that may contribute to AF risk.
Methods
Our study was conducted using large population-based cohort studies comprised of both Black and White individuals that had systematically ascertained incident AF and whose participants had undergone genome wide analysis of SNPs using DNA microarrays. The design, recruitment, baseline characterization, and outcome ascertainment procedures for these cohorts have been previously described in detail and brief summaries are provided below.22–25
Study Cohorts
Cardiovascular Health Study (CHS)
CHS enrolled a total of 5,201 participants aged 65 years and older that were recruited in 1989–1990 from Medicare eligibility lists in four US communities: Forsyth County, North Carolina; Washington County, Maryland; Sacramento County, California; and Pittsburgh, Pennsylvania. An additional 687 participants, almost all Black, were recruited into the study in 1992–1993. Participants underwent comprehensive examinations at study entry to document baseline demographics and medical co-morbidities. Methods for ascertaining prevalent hypertension, diabetes, heart failure, and coronary artery disease have been previously described.23 Subsequent follow-up was performed with alternating clinic visits and phone calls every six months until 1999 and semi-annual phone calls thereafter. Resting 12-lead ECGs were performed at each clinic visit. Prevalent AF at baseline was documented using the baseline ECG, while incident AF was ascertained on the basis of clinic visit ECGs and hospital discharge diagnosis codes that were supplemented with Medicare inpatient and outpatient claims data. 26
Atherosclerosis Risk in Communities Study (ARIC)
ARIC enrolled 15,792 adults aged 45–64 years between 1987 and 1989 from 4 US communities: the northwest suburbs of Minneapolis, MN; Washington County, MD; Jackson, MS; and Forsyth County, NC. Comprehensive baseline evaluations were performed followed by annual phone interviews and 3 repeat examinations spaced approximately 3 years apart. Methods for ascertaining baseline co-morbidities including hypertension, diabetes, heart failure, and coronary artery disease have been documented previously.24 Standard resting 12-lead ECGs were performed at each visit. Prevalent AF was identified from the baseline ECG, and incident AF was identified from study visit ECGs, hospital discharge diagnoses, and death certificates.6
Health ABC
Health ABC enrolled 3,075 adults aged 70–79 years between 1997 and 1998 from 2 US communities: Pittsburgh, PA and Memphis, TN. Participants underwent a comprehensive baseline examination followed by clinic visits at 2, 3, 4, 5, 6, 8, and 10 years with alternating biannual phone calls. Phone calls continued through Year 15 of the study. Methods for ascertaining prevalent hypertension, diabetes, heart failure, and coronary artery disease have been previously described.25 Standard resting 12-lead ECGs were performed at the baseline and 4 year visits. Prevalent AF was identified using the baseline ECG and Medicare claims data. Incident AF was identified from the fourth year ECG, hospital discharge diagnoses, or Medicare claims data.27
Genotyping, SNP Selection, and Imputation
Genotyping of study participants was completed using DNA microarrays or chips that permit simultaneous analysis of hundreds of thousands of SNPs present throughout the genome. CHS performed whole genome SNP analysis using the Illumina 370 CNV and HumanOmni1-Quad_v1 BeadChip system in Whites and Blacks, respectively (Illumina, San Diego, CA). DNA analysis in Health ABC was performed using the Illumina Human 1M-Duo microarray. Variant calling for both studies was performed using the Illumina BeadStudio algorithm. Genotyping within ARIC was performed on the Affymetrix 6.0 DNA microarray (Affymetrix, Santa Clara, CA) and analyzed with the Birdseed variant calling algorithm.13 Although different genotyping platforms were utilized in the different cohorts, comparison of measures of associations between cohorts was still considered valid, consistent with the approach utilized in other genetic studies.13 Quality control criteria were subsequently utilized to filter SNPs containing possible errors. SNPs were excluded from analysis based on previously established thresholds within each cohort for SNP call rates, Hardy-Weinberg p-values, and minor allele frequencies (MAFs).13 Individuals with either known or cryptic relatedness were also excluded.
Analysis of the 9 AF-risk SNPs was performed in both the CHS and ARIC cohorts. Three of the 9 SNPs (rs2200733, rs3807989, rs10824026) were directly genotyped in CHS, while 2 (rs2200733, rs2106261) were directly genotyped in ARIC. The remaining SNPs were imputed after phasing the datasets with SHAPEIT.28 Imputation serves as a highly accurate method to determine SNP carrier status when they were not directly genotyped as part of genome wide SNP analysis. For each SNP, a surrounding 1 Megabase region was selected and imputed using IMPUTE2 with the full 1000 Genomes dataset (Phase 1 v3) as the reference.29 For each of the SNPs, we checked the imputation quality and found it to be high in all of the datasets (IMPUTE2 info score > 0.95) where sufficient reference overlap was present to perform imputation.
Polygenic Risk Score
Polygenic risk scores based on the 9 AF-associated SNPs were calculated for both Whites and Blacks in the CHS and ARIC cohorts. The polygenic risk score was based on the odds ratio determined for each SNP in the prior AF GWAS meta-analysis among Whites.13 The odds ratio for each SNP was exponentiated to the number of risk alleles carried by each participant and the overall polygenic risk score was the product of these 9 values.30 This calculation assumes a multiplicative risk across loci with no interaction terms.
Locus Specific Ancestry Estimation for Admixture Mapping
Locus-specific African ancestry across the genome in Blacks was estimated using the program LAMPLD 1.0 using a 2 population model (African and European).31 Genotype data for Yoruba in Ibadan, Nigeria and Utah Residents with Northern and Western European Ancestry from The International HapMap Project (www.hapmap.org) were used as ancestral reference data for the 2 ancestral populations. The use of Yoruban genotypes was based on previous work indicating that Yorubans serve as the best reflection of African ancestry among modern day Africans.32 The mean of locus-specific African ancestry across the genome was calculated and used as the global African ancestry for each Black individual. The analysis included 596,464 markers from ARIC, 588,058 markers from CHS, and 876,739 markers from Health ABC.
Statistical Analysis
Normally distributed continuous variables are presented as means ± standard deviation and were compared using the Student’s t-test. Comparison of categorical values and minor allele frequencies for Blacks and Whites was performed using the Chi-squared test.
Time-to-event analyses using Cox proportional hazards models were employed to evaluate for an association between each of the 9 SNPs and incident AF. Covariates included in these models were baseline age, gender, body mass index, hypertension, diabetes, heart failure and coronary artery disease. Associations between polygenic risk scores and incident AF were also determined using Cox proportional hazards models after controlling for the aforementioned covariates. The proportion of treatment effect (PTE) method was utilized to determine if SNP carrier status mediated the association between race and AF through time-to-event analyses using adjusted Cox proportional hazards models and the previously specified covariates.33 The PTE evaluates the magnitude of effect of a putative mediator based on the proportional reduction in the regression coefficient of the predictor of interest when the putative mediator is included in the Cox proportional hazards model (a more detailed description of the PTE is provided in the Online Supplement, eMethod2). The impact of each SNP was examined in isolation, while a model containing all SNPs also examined their cumulative effect. Confidence intervals were obtained using bootstrap resampling with 1,000 repetitions.
Cox proportional hazards models were also employed to evaluate for an association between locus-specific African ancestry and incident AF in the admixture mapping component of the study. Locus specific ancestry was coded as a continuous variable between 0 and 2 where 0 was equivalent to having 0% probability of any African chromosomes and 2 is equivalent to 100% probability of having 2 African chromosomes at a locus. In order to adjust for potential confounding, covariates added to these models included baseline age, gender, body mass index, hypertension, diabetes, left ventricular hypertrophy, prevalent heart failure, history of myocardial infarction, study site, and % genome-wide African ancestry for each individual (% European ancestry is accounted for by the % African ancestry in the model since the two are, by definition of the model, forced to add up to 100%).
Given previous evidence that racial differences in AF may be most pronounced among those without conventional AF risk factors12, the admixture mapping was performed using all AF as the primary outcome and AF in the absence of pre-specified baseline risk factors as a secondary outcome. For the second analysis, participants with prevalent hypertension, diabetes, heart failure, and history of myocardial infarction at baseline were excluded. Covariates included in the multivariate Cox proportional hazards models were otherwise the same as above. Meta-analyses with inverse variance weighting were performed on the results from all 3 studies for both the overall cohorts and the lone AF analyses using the program METAL.34 The genome-wide significance threshold for admixture mapping was 7×10−6, which corresponds to a Bonferroni correction for testing 7000 independent ancestry blocks in the genome and was derived empirically based on permutations.35
Admixture mapping was performed using the survival analysis package in R. The remainder of the statistical analyses were performed using Stata version 12 (College Station, Tx, USA). With the exception of the genome wide admixture mapping analyses, two-tailed p-values < 0.05 were considered statistically significant.
Results
Study Participants
CHS
A total of 3,362 (84.7%) individuals from the CHS cohort had a self-reported race of White, while 811 were Black. The baseline clinical characteristics of the cohort are summarized in Table 1. During a mean follow-up period of 11.2 years, a total of 1,023 individuals were diagnosed with incident AF. Among the overall cohort, a subgroup of 589 participants were classified as having developed AF in the absence of the pre-specified risk factors described above.
Table 1.
Baseline Characteristics of CHS, ARIC, and HABC White and Black Study Participants
| CHS | ARIC | Health ABC | |||
|---|---|---|---|---|---|
| Whites N = 3362 |
Black N = 811 |
Whites N = 9229 |
Blacks N = 3112 |
Blacks N = 1015 |
|
| Age (years) | 72.3 ± 5.4 | 72.7 ± 5.6 | 54.3 ± 5.7 | 53.3 ± 5.8 | 73.4 ± 2.9 |
| Male (%) | 40.0 | 36.6 | 47.1 | 37.4 | 42.8 |
| Hypertension (%) | 31.8 | 54.8 | 26.8 | 55.5 | 70.3 |
| Diabetes Mellitus (%) | 12.1 | 21.6 | 8.7 | 19.8 | 20.8 |
| Body Mass Index (kg/m2) | 26.3 ± 4.4 | 28.4 ± 5.3 | 27.0 ± 4.8 | 29.7 ± 6.1 | 28.5 ± 5.3 |
| Coronary Artery Disease (%) | 2.0 | 3.5 | 5.1 | 4.0 | 20.1 |
| Congestive Heart Failure (%) | 0.51 | 0.99 | 3.8 | 6.6 | 2.6 |
Data are n (%) or mean ± standard deviation, CHS = Cardiovascular Healthy Study, ARIC = Atherosclerosis Risk in Communities Study, Health ABC = Health, Aging, and Body Composition Study
ARIC
Within the ARIC cohort, 9,229 (74.8%) and 3,112 reported their race as being White and Black, respectively. Baseline clinical characteristics of the cohort are summarized in Table 1. Among 12,341 individuals, a total of 1,341 were diagnosed with incident AF during a mean follow-up period of 17.5 years and a subgroup of 569 participants developed incident AF in the absence of the pre-specified AF risk factors at baseline.
Health ABC
A total of 1,078 individuals from the Health ABC cohort had a self-reported race of Black and underwent genome wide SNP analysis. Among this group, 63 had prevalent AF and were excluded from the analysis. The remaining baseline clinical characteristics of the cohort are summarized in Table 1. During a mean follow-up period of 10.5 years, a total of 237 individuals were diagnosed with incident AF. Among the overall cohort, a subgroup of 24 participants developed incident AF in the absence of the baseline pre-specified risk factors.
Impact of 9 AF-Risk SNPs on the Differential Racial Risk of AF
We found significant differences in the allele frequencies between races for each of the SNPs (all p < 0.001) (Table 2). The direction and magnitude of the MAF differences between races were consistent in both cohorts. The 3 SNPs with the strongest associations with AF in prior studies (rs2200733, rs2106261, and rs6666258) had markedly greater risk allele frequencies among Blacks relative to Whites (Table 2).
Table 2.
Minor Allele Frequencies of Atrial Fibrillation Associated SNPs in CHS and ARIC Among Whites and Blacks
| SNP | Genetic Locus | Nearest Gene | Impact of Minor Allele on AF Risk | CHS | p value | ARIC | p value | ||
|---|---|---|---|---|---|---|---|---|---|
| White n = 3362 |
Black n = 811 |
White n = 9229 |
Black n = 3112 |
||||||
| rs2200733 | 4q25 | PITX2 | Increased | 13.7 | 20.3 | <0.001 | 11.5 | 21.6 | <0.001 |
| rs2106261 | 16q22 | ZFHX3 | Increased | 16.4 | 26.4 | <0.001 | 17.0 | 26.1 | <0.001 |
| rs6666258 | 1q21 | KCNN3 | Increased | 28.7 | 35.7 | <0.001 | 30.3 | 36.1 | <0.001 |
| rs3903239 | 1q24 | PRRX1 | Increased | 42.6 | 21.3 | <0.001 | 43.8 | 18.6 | <0.001 |
| rs3807989 | 7q31 | CAV1 | Protective | 40.3 | 36.8 | 0.083 | 40.8 | 36.2 | <0.001 |
| rs10821415 | 9q22 | C9orf3 | Increased | 42.4 | 21.7 | <0.001 | 42.2 | 20.6 | <0.001 |
| rs10824026 | 10q22 | SYNPO2L | Protective | 16.4 | 37.7 | <0.001 | 15.5 | 37.8 | <0.001 |
| rs1152591 | 14q23 | SYNE2 | Increased | 46.5 | 14.4 | <0.001 | 48.1 | 12.8 | <0.001 |
| rs7164883 | 15q24 | HCN4 | Increased | 16.9 | 32.1 | <0.001 | 15.6 | 32.7 | <0.001 |
Minor Allele Frequencies are reported as %. SNP = single nucleotide polymorphism, CHS = Cardiovascular Health Study, ARIC = Atherosclerosis Research in Communities Studies
We evaluated the impact of each SNP on AF risk in both the CHS and ARIC cohorts for both Whites and Blacks. Findings were generally consistent between the cohorts, races, and the previously reported results (Online Supplement, eTable 1). The polygenic risk scores predicted incident AF among Whites and Blacks in both cohorts (Online Supplement, eTable 2).
We used the PTE to evaluate the effect of these SNPs on the differential racial risk of the arrhythmia. In an overall model containing each SNP, there was evidence for mediation of the race-AF relationship in ARIC (PTE: 24.8; 95% CI: 3.0, 49.9, p=0.03), however this was not replicated in CHS (PTE: −12.4; 95% CI: −67.7, 13.4, p=0.74). One SNP, rs10824026 (chromosome 10: position 73661450; intronic within SYNPO2L), appeared to be a mediator of the relationship in the anticipated direction consistent with a protective effect of the SNP on the risk of AF in both CHS and ARIC (Table 3). The rs10824026 SNP was estimated to account for 11.4% (95% CI 2.9–29.9, p=0.01) and 31.7% (95% CI 16.0–53.0, p<0.01) of the increased risk of the arrhythmia in Whites relative to Blacks in CHS and ARIC, respectively. None of the remaining 8 SNPs exhibited statistically significant mediation effects in the anticipated direction in both cohorts (Table 3). The allele previously shown to exhibit a protective effect against AF, was more common among Blacks than Whites in both CHS and ARIC (Table 2). The MAF among Blacks ranged from 37.7 – 37.8% in comparison with 15.5–16.4% among Whites.
Table 3.
Proportion of Treatment Effect of Atrial Fibrillation Associated Single Nucleotide Polymorphisms on the Differential Risk of the Arrhythmia Among Whites and Blacks
| SNP | Genetic Locus | Nearest Gene | CHS | ARIC | ||||
|---|---|---|---|---|---|---|---|---|
| PTE | 95% CI | p-value | PTE | 95% CI | p-value | |||
| rs2200733 | 4q25 | PITX2 | −4.58 | −16.82, −0.78 | 0.03 | −11.7 | −18.24, −6.35 | <0.01 |
| rs2106261 | 16q22 | ZFHX3 | −0.84 | −5.24, 1.28 | 0.54 | −4.91 | −8.36, −1.72 | <0.01 |
| rs6666258 | 1q21 | KCNN3 | −3.14 | −14.19, 0.98 | 0.14 | −2.60 | −5.05, −0.68 | <0.01 |
| rs3903239 | 1q24 | PRRX1 | −1.70 | −7.73, 2.18 | 0.34 | 6.73 | 0.46, 13.52 | 0.03 |
| rs3807989 | 7q31 | CAV1 | −1.61 | −7.11, 1.00 | 0.31 | −2.36 | −8.19, 2.95 | 0.38 |
| rs10821415 | 9q22 | C9orf3 | −4.52 | −16.22, 2.44 | 0.21 | 5.14 | −1.52, 11.27 | 0.11 |
| rs10824026 | 10q22 | SYNPO2L | 11.40 | 2.90, 29.87 | 0.01 | 28.55 | 13.62, 47.23 | <0.01 |
| rs1152591 | 14q23 | SYNE2 | 9.17 | −0.09, 29.35 | 0.07 | 12.99 | 2.96, 24.54 | <0.01 |
| rs7164883 | 15q24 | HCN4 | −4.52 | −14.98, 0.81 | 0.11 | −8.03 | −14.78, −3.00 | <0.01 |
SNP = Single Nucleotide Polymorphism, CHS = Cardiovascular Health Study, ARIC = Atherosclerosis Research in Communities Studies, PTE = Proportion of Treatment Effect, CI = Confidence Interval
Admixture Mapping
Since the majority of the risk difference between the groups was unexplained by the 9 known risk SNPs, we searched for new loci that may affect AF risk using admixture mapping. In order to enhance the power, data from a third cohort, Health ABC, was added.
Meta-Analysis
The results of admixture mapping from the 3 individual cohorts are provided in the eAppendix in the Supplement. Combined analysis of the 3 cohorts identified the peak for the locus specific ancestry association at rs1977904 (chr 21: position 31589767) (Figure 1D). In contrast to the top loci in the individual cohorts, this locus conferred an increased risk of incident AF in association with African ancestry (HR 1.42, 95% CI: 1.20–1.67, p=2.40 × 10−5) for each additional African chromosome. Among the pre-specified AF subgroup lacking baseline risk factors, the strongest locus specific ancestry association was at rs4649274 (chr 1: position 230801857) (Figure 2D). Unlike the overall meta-analysis, but consistent with the observed differential racial risk of the arrhythmia, African ancestry was associated with a reduced likelihood of incident AF within the pre-specified AF subgroup (HR: 0.49 95% CI: 0.35–0.69, p=4.44 × 10−5). The peak locus specific ancestry associations in the meta-analyses for both the overall cohort and the pre-specified AF subgroup failed to meet the threshold for genome-wide significance (p=7×10−6).
Figure 1.
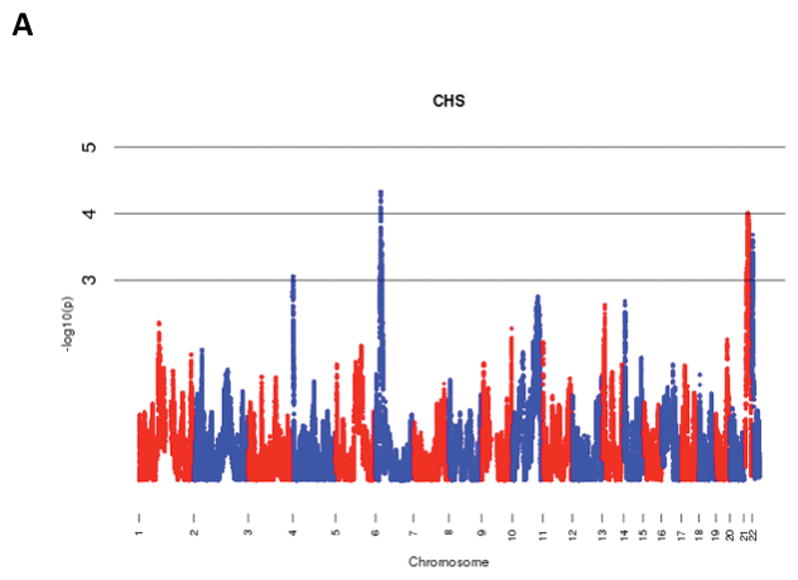
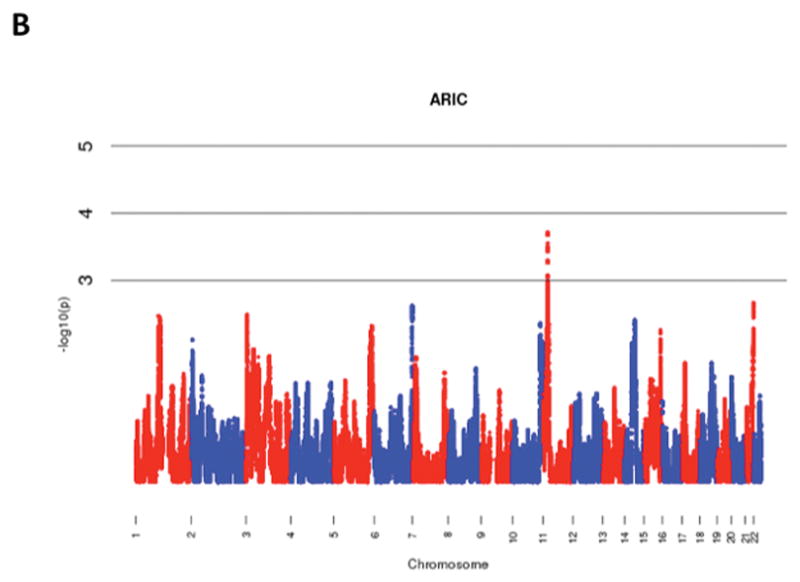
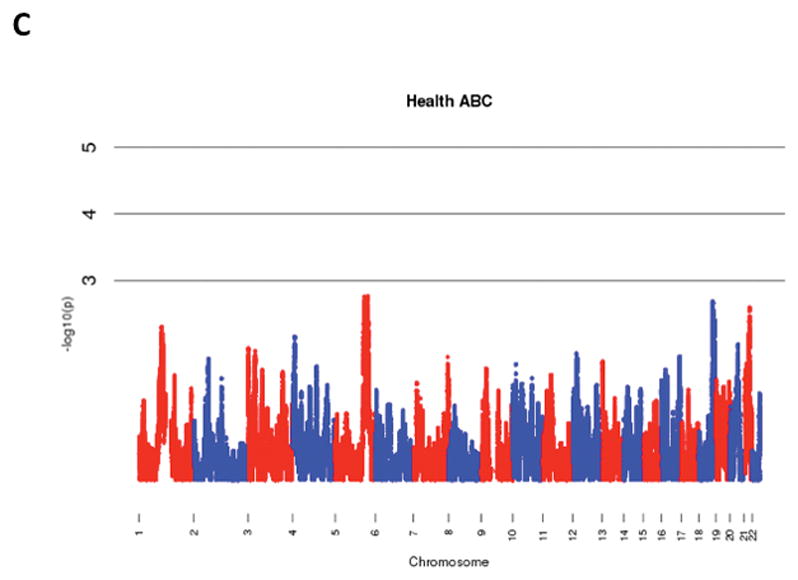
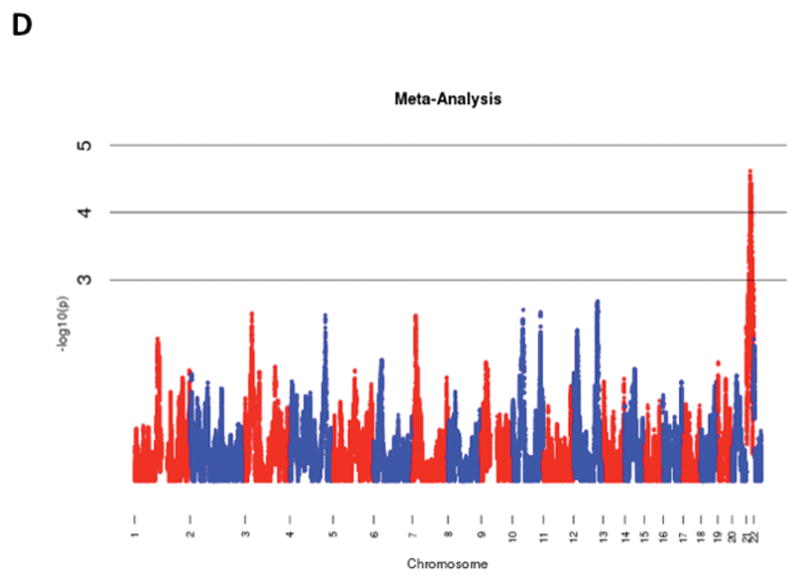
Manhattan Plots of Admixture Mapping for Incident Atrial Fibrillation Among Blacks from CHS (A), ARIC (B), Health ABC (C), and meta-analysis (D).
Figure 2.
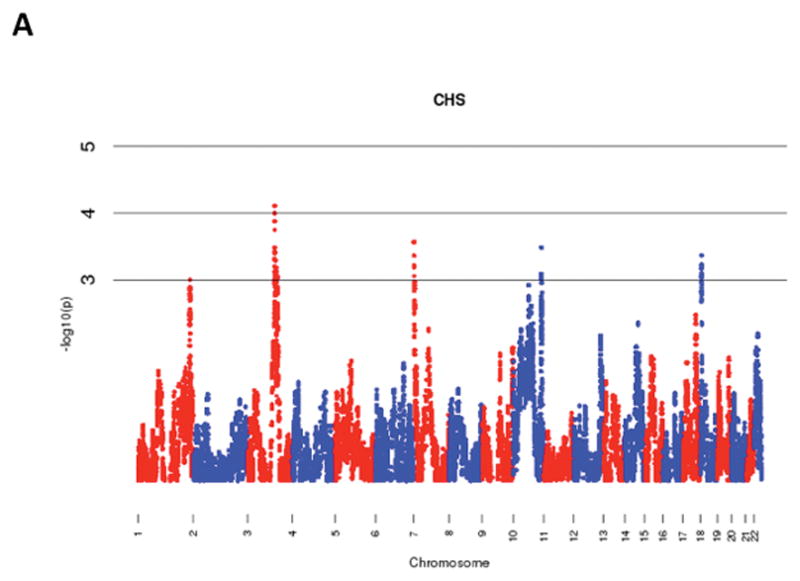
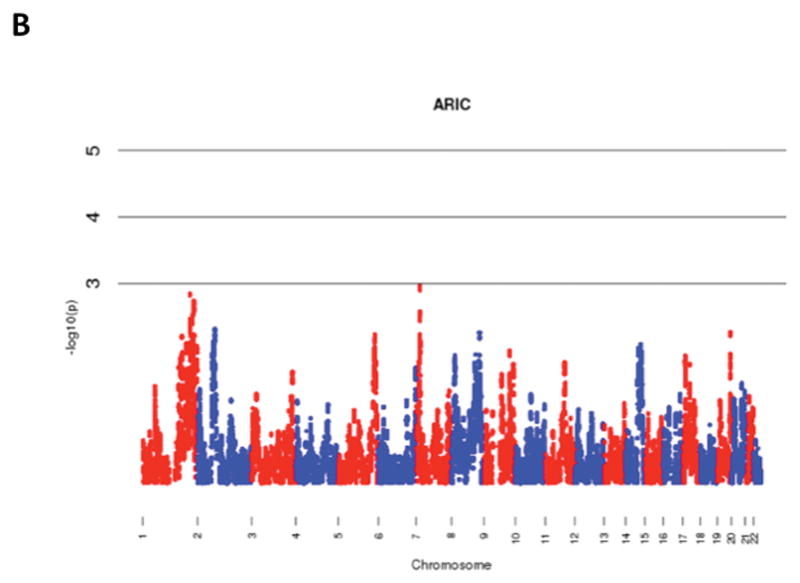
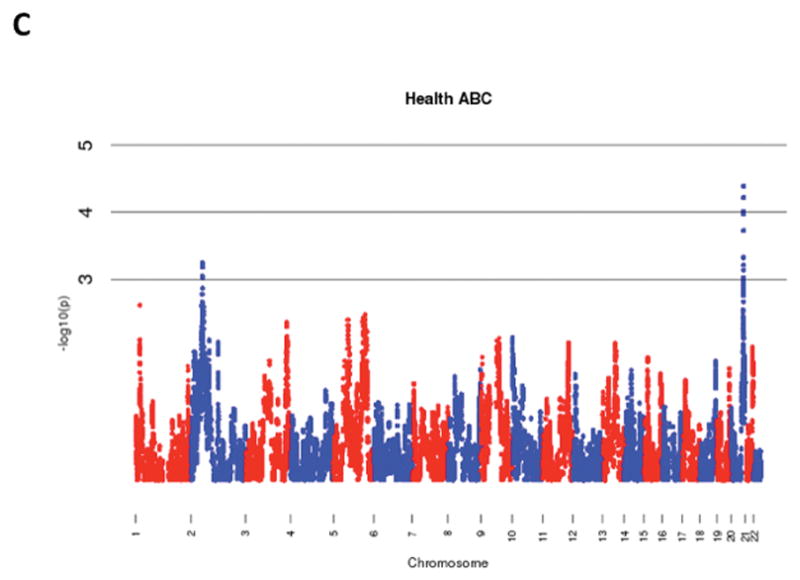
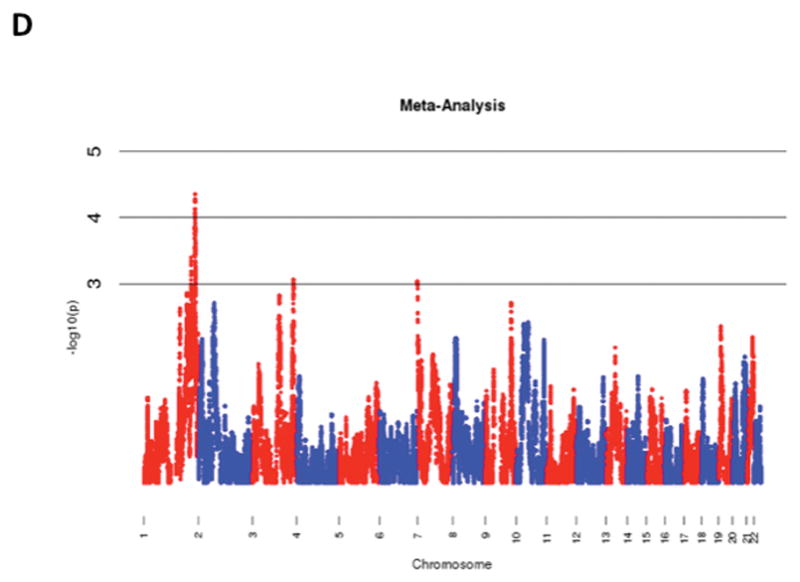
Manhattan Plots of Admixture Mapping Restricted to Incident Cases of Atrial Fibrillation Among Blacks that Developed the Arrhythmia in the Absence of Baseline Pre-Specified Risk Factors* from CHS (A), ARIC (B), Health ABC (C), and meta-analysis (D).
*Hypertension, Diabetes, Heart failure, and History of Myocardial Infarction
Admixture Mapping and rs10824026
The impact of the locus specific ancestry associated with rs10824026, the SNP found to partially mediate the increased risk of AF among Whites relative to Blacks in the initial analysis, was also examined in the admixture mapping study. Because it was not directly genotyped, the SNP in greatest linkage disequilibrium with rs10824026 (rs12570126; r2 = 0.935, D′ = 1.00) was used as a proxy to estimate its locus specific ancestry association. In the overall meta-analysis, the locus specific ancestry associated with the rs12570126 SNP exhibited a non-significant lower risk of AF among Blacks (HR: 0.78, 95% CI: 0.54–1.12, p=0.174).
Discussion
Our investigation into the genetics underlying the differential racial risk of AF suggests that one of the SNPs with an established association with AF, rs10824026, may partially mediate the increased risk of the arrhythmia among Whites relative to Blacks. The minor allele of the SNP is known to be protective against AF and is significantly more common among Blacks than Whites. Our study suggests that the rs10824026 SNP may account for 11.4% to 31.7% of the reduced risk of AF observed among Blacks relative to Whites. The admixture mapping component of our study involving 3 large population-based cohorts failed to identify a novel genetic variant that reached genome wide significance on meta-analysis emphasizing that a single SNP in isolation is unlikely to account for the paradoxical differential racial risk of AF. Within the subgroup analysis of individuals that developed AF in the absence of pre-specified baseline risk factors, however, the top locus with non-significant association by admixture mapping conferred an increased risk of incident AF in association with European ancestry consistent with the known epidemiology.
The rs10824026 SNP, present within the 10q22 genetic locus, is located 5 and 20 kb upstream of the SYNPO2L and MYOZ1 genes, respectively. Both of these genes are expressed within cardiac tissue, however their precise functions are unknown. SYNPO2L encodes the synaptopodin 2-like protein and, as suggested, has structural similarities with the SYNPO2 gene, whose name is derived from its presence within renal podocytes and post-synaptic densities and dendritic spines. Review of the expression Quantitative Trait Loci (eQTL) Genotype-Tissue Expression (GTEx) database (http://www.gtexportal.org/home/) suggested that, although the SNP is in closer proximity to SYNPO2L, its biological effect is actually mediated through altered expression levels of MYOZ1.36 GTEx is an eQTL database that possesses genome wide SNP and RNA transcript data that permits evaluation of SNP status in relation to transcript levels of nearby genes in specific tissues. In analyses restricted to atrial tissue, the rs10824026 SNP was associated with a significant change in transcript levels of a single gene, MYOZ1 (Effect Size = −1.1, p = 4.0e-16). The myozenin-1 protein, encoded by MYOZ1, appears to influence calcineurin signaling and interacts with proteins at the Z-disc of the cardiac sarcomere, including α-actinin and γ-filamin.37 These eQTL findings are consistent with previous work suggesting that the AF risk residing in the chromosome 10q22 locus is secondary to MYOZ1.38
The finding that the rs10824026 SNP mediates the reduced risk of AF observed among Blacks relative to Whites is consistent with the minor allele being protective against AF and more common in Blacks. In contrast, it is interesting to note that the MAFs of the 3 SNPs with the strongest associations with AF (rs2200733, rs2106261, and rs6666258) are markedly more common in Blacks relative to Whites (Table 2). This finding is especially surprising given that the minor allele of each of these 3 SNPs is associated with an increased risk of the arrhythmia among both races and hence would be anticipated to be more common among Whites. Although an explanation for these observations is not immediately clear, the lower risk of the arrhythmia within Blacks is likely secondary to additional genetic and/or environmental factors that serve to at least partially counteract the effect of these SNPs.
Within the admixture mapping component of the study, there were no loci that met the pre-specified threshold for genome wide significance. In contrast to the overall meta-analysis, the top hit within the subgroup analysis involving individuals that developed AF in the absence of the pre-specified risk factors was associated in the anticipated direction, consistent with a reduced risk of the arrhythmia among Blacks relative to Whites. The SNP with the strongest locus specific ancestry association in this subgroup analysis, rs4649274, is intronic within SIPA1L2, a gene located on chromosome 1 that encodes the signal-induced proliferation-associated 1 like 2 protein. The function of SIPA1L2 is currently unclear, however it is expressed in the heart and could potentially exert a role in AF pathophysiology. The gene from which it derives its name, SIPA1, encodes a mitogen induced GTPase activating protein.
Our inability to identify SNPs responsible for the differential racial risk of AF with admixture mapping highlights the increasing difficulty of uncovering novel genetic variants that predispose to the arrhythmia. The majority of the heritability of the arrhythmia remains unexplained and may be secondary to multiple weak signals that fall below our current level of detection, gene-gene and gene-environmental interactions, and/or environmental exposures in isolation.39 It is also possible that the heritability of the arrhythmia is primarily mediated by rare variants and large-scale whole exome and genome initiatives will be required to uncover the remaining genetic culprits.
Our study has several potential limitations. As discussed, the failure to identify a statistically significant genetic variant with admixture mapping may be secondary to inadequate power given that our study only comprised a total of 4,938 Blacks in the overall meta-analysis. It should be noted, however, that these are the 3 largest population-based cohort studies involving Blacks that have undergone GWAS analysis and consequently our study leverages the most extensive data available. Second, these cohorts, particularly CHS and Health ABC, are predominantly comprised of older individuals. The impact of genetics on AF risk is generally more modest in older age groups and therefore our results may not be generalizable to younger populations. It should also be noted that we did not adjust for multiple hypothesis testing in the analysis of the 9 previously established AF-associated SNPs. Although increasing the possibility of a spurious association, the involvement of these 9 SNPs with AF has been firmly established and they were identified a priori for this analysis. In addition, we further minimized the possibility of a type 1 error through replication in a second cohort. Multiple SNPs in our candidate SNP analysis were not directly genotyped, but instead were imputed (each with high probability; >0.95). Although imputation may introduce some error, it is anticipated to be random. Any resulting bias would be expected to occur towards the null and therefore should not lead to spurious false positive associations. Lastly, there have been 5 additional SNPs found to be associated with AF beyond the 9 evaluated in this study.14 In order to try to minimize the impact of multiple hypothesis testing, we did not examine for a role of these additional SNPs, however their potential involvement as mediators should be evaluated in future studies.
Conclusions
Our investigation into the genetic determinants accounting for the paradoxical differential racial risk of AF among Whites and Blacks suggests that a previously known AF-associated SNP, rs10824026, may be a partial mediator of this relationship. The admixture mapping component of our analysis failed to identify any novel genetic variants. Taken together, these findings demonstrate that additional genetic or environmental influences beyond single SNPs in isolation are likely operational. Given the consistent observations that race is a powerful risk factor for AF that can “trump” conventional risk factors5–8, our data suggest that this phenomenon is primarily driven by either polygenic influences, gene-environment interactions, or environmental influences in isolation.
Supplementary Material
Acknowledgments
The funders of this research, the American Heart Association and the Joseph Drown Foundation, had no role in the design and conduct of the study; no role in the collection, management, analysis, and interpretation of the data; no role in the preparation, review, or approval of the manuscript; and no role in the decision to submit the manuscript for publication. Dr. Marcus had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis
Funding Sources: This work was made possible by an American Heart Association Western States Afilliate Grant (G.M.M) and the Joseph Drown Foundation (G.M.M.). J.D.R. is supported by research fellowship grants from the Heart & Stroke Foundation of Canada and the Canadian Heart Rhythm Society. Grants for the Cardiovascular Health Study include: R21 HL121348, 1R01 HL127659, R01 HL80698-01, NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01 HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and grants HL080295 and HL102214, with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided by AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. Research involving the Health, Aging, and Body Composition (Health ABC) study was supported by NIA contracts N01AG62101, N01AG62103, and N01AG62106 and, in part, by the INTRAMURAL RESEARCH PROGRAM of the NIA of the NIH. The genome-wide association study was funded by NIA grant 1R01AG032098- 01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosures: None to report and no relevant relationships with industry.
References
- 1.Wang TJ, Larson MG, Levy D, et al. Temporal relations of atrial fibrillation and congestive heart failure and their joint influence on mortality: the Framingham Heart Study. Circulation. 2003;107(23):2920–2925. doi: 10.1161/01.CIR.0000072767.89944.6E. [DOI] [PubMed] [Google Scholar]
- 2.Wolf PA, Abbott RD, Kannel WB. Atrial fibrillation as an independent risk factor for stroke: the Framingham Study. Stroke. 1991;22(8):983–988. doi: 10.1161/01.str.22.8.983. [DOI] [PubMed] [Google Scholar]
- 3.Benjamin EJ, Wolf PA, D’Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation. 1998;98(10):946–952. doi: 10.1161/01.cir.98.10.946. [DOI] [PubMed] [Google Scholar]
- 4.Psaty BM, Manolio TA, Kuller LH, et al. Incidence of and risk factors for atrial fibrillation in older adults. Circulation. 1997;96(7):2455–2461. doi: 10.1161/01.cir.96.7.2455. [DOI] [PubMed] [Google Scholar]
- 5.Dewland TA, Olgin JE, Vittinghoff E, Marcus GM. Incident atrial fibrillation among Asians, Hispanics, blacks, and whites. Circulation. 2013;128(23):2470–2477. doi: 10.1161/CIRCULATIONAHA.113.002449. [DOI] [PubMed] [Google Scholar]
- 6.Alonso A, Agarwal SK, Soliman EZ, et al. Incidence of atrial fibrillation in whites and African-Americans: the Atherosclerosis Risk in Communities (ARIC) study. Am Heart J. 2009;158(1):111–117. doi: 10.1016/j.ahj.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcus GM, Olgin JE, Whooley M, et al. Racial differences in atrial fibrillation prevalence and left atrial size. Am J Med. 2010;123(4):375, e1–e7. doi: 10.1016/j.amjmed.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcus GM, Alonso A, Peralta CA, et al. European ancestry as a risk factor for atrial fibrillation in African Americans. Circulation. 2010;122(20):2009–2015. doi: 10.1161/CIRCULATIONAHA.110.958306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448(7151):353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 10.Gudbjartsson DF, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41(8):876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benjamin EJ, Rice KM, Arking DE, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41(8):879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellinor PT, Lunetta KL, Glazer NL, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42(3):240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellinor PT, Lunetta KL, Albert CM, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44(6):670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinner MF, Tucker NR, Lunetta KL, et al. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation. 2014;130(15):1225–1235. doi: 10.1161/CIRCULATIONAHA.114.009892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopp JB, Smith MW, Nelson GW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40(10):1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genovese G, Friedman DJ, Ross MD, et al. Association of Trypanolytic ApoL1 Variants with Kidney Disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103(38):14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nalls MA, Wilson JG, Patterson NJ, et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am J Hum Genet. 2008;82(1):81–87. doi: 10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reich D, Nalls MA, Kao WHL, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5(1):e1000360. doi: 10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fejerman L, Ahmadiyeh N, Hu D, et al. Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nat Commun. 2014;5:5260. doi: 10.1038/ncomms6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deo RC, Wilson JG, Xing C, et al. Single-nucleotide polymorphisms in LPA explain most of the ancestry-specific variation in Lp(a) levels in African Americans. PloS One. 2011;6(1):e14581. doi: 10.1371/journal.pone.0014581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1(3):263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 23.Tell GS, Fried LP, Hermanson B, Manolio TA, Newman AB, Borhani NO. Recruitment of adults 65 years and older as participants in the Cardiovascular Health Study. Ann Epidemiol. 1993;3(4):358–366. doi: 10.1016/1047-2797(93)90062-9. [DOI] [PubMed] [Google Scholar]
- 24.Investigators TA. The Atherosclerosis Risk in Communit (aric) Stui)y: Design and Objectwes. Am J Epidemiol. 1989;129(4):687–702. [PubMed] [Google Scholar]
- 25.Newman AB, Simonsick EM, Naydeck BL, et al. Association of long-distance corridor walk performance with mortality, cardiovascular disease, mobility limitation, and disability. JAMA. 2006;295(17):2018–2026. doi: 10.1001/jama.295.17.2018. [DOI] [PubMed] [Google Scholar]
- 26.Ives DG, Fitzpatrick AL, Bild DE, et al. Surveillance and ascertainment of cardiovascular events. The Cardiovascular Health Study. Ann Epidemiol. 1995;5(4):278–285. doi: 10.1016/1047-2797(94)00093-9. [DOI] [PubMed] [Google Scholar]
- 27.Auer R, Bauer DC, Marques-Vidal P, et al. Association of major and minor ECG abnormalities with coronary heart disease events. JAMA. 2012;307(14):1497–1505. doi: 10.1001/jama.2012.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 29.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vachon CM, Pankratz VS, Scott CG, et al. The contributions of breast density and common genetic variation to breast cancer risk. J Natl Cancer Inst. 2015;107(5) doi: 10.1093/jnci/dju397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baran Y, Pasaniuc B, Sankararaman S, et al. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics. 2012;28(10):1359–1367. doi: 10.1093/bioinformatics/bts144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zakharia F, Basu A, Absher D, et al. Characterizing the admixed African ancestry of African Americans. Genome Biol. 2009;10(12):R141. doi: 10.1186/gb-2009-10-12-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin DY, Fleming TR, De Gruttola V. Estimating the proportion of treatment effect explained by a surrogate marker. Stat Med. 1997;16(13):1515–1527. doi: 10.1002/(sici)1097-0258(19970715)16:13<1515::aid-sim572>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 34.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang H, Siegmund DO, Johnson NA, Romieu I, London SJ. Joint testing of genotype and ancestry association in admixed families. Genet Epidemiol. 2010;34(8):783–791. doi: 10.1002/gepi.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takada F, Woude DLV, Tong H-Q, et al. Myozenin: An α-actinin- and γ-filamin-binding protein of skeletal muscle Z lines. Proc Natl Acad Sci. 2001;98(4):1595–1600. doi: 10.1073/pnas.041609698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin H, Dolmatova EV, Morley MP, et al. Gene Expression and Genetic Variation in Human Atria. Heart Rhythm. 2014;11(2):266–271. doi: 10.1016/j.hrthm.2013.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y, Wang C, Yao Y, et al. Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation. PLoS Genet. 2015;11(8):e1005393. doi: 10.1371/journal.pgen.1005393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


