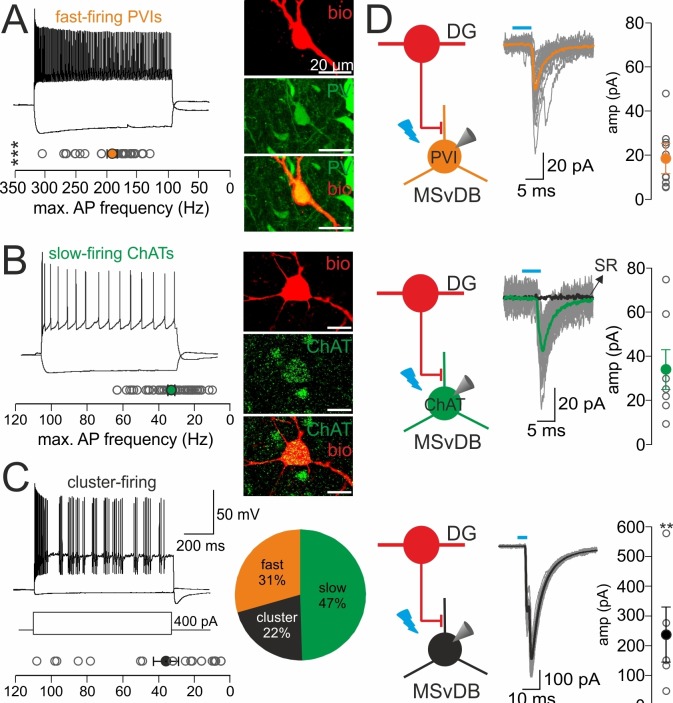
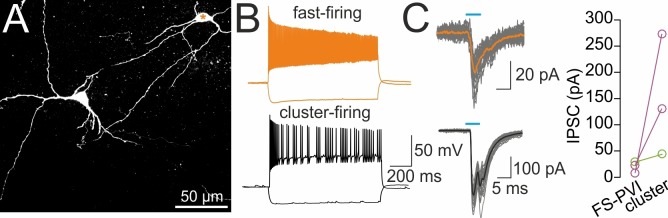
Figure 6. DG-SOMIs provide weak inhibition on GABAergic and cholinergic but strong inhibition onto putative glutamatergic cells in the medial septum.
(A–C) Passive and active membrane properties and the neurochemical marker contents of the three main neuron types in the medial septum and vertical limb of the diagonal band of Broca (MSvDB). Left, characteristic discharge patterns of the three cells types (1 s, −100 and 400 pA current injection) classify them as fast-spiking (A), slow-firing (B) and cluster-firing (C) cells. Plots summarize the maximal discharge frequency of the three cell types. The discharge frequency between PVIs and slow-firing as well as cluster-firing cells was significantly different (***p<0.001 for both comparisons, t-test). Right, intracellular labeling of cells with biocytin (red) with subsequent antibody labeling (green). Note, all fast-firing cells expressed parvalbumin (PV). Slow-firing cells express choline acetyltransferase (ChAT) and cluster-firing cells have been previously identified as glutamatergic (Glut) cells (Manseau et al., 2005; Mattis et al., 2014). Pie chart summarizes the relative proportion of the recorded cell types (32 fast-spiking PVIs, 49 slow-firing ChATs, 23 cluster-firing putative Glut cells). (D) Left, schematic illustration of the experimental procedure. DG-septal projecting SOMIs expressing channelrhodopsin-2 (ChR2) after injection of rAAV-FLEX-ChR2-tdT in the dorsal DG were activated by light-pulses (5 ms, full-field illumination, 473 nm) applied to the MSvDB (red circles represent HIL cells). Middle, IPSCs recorded in the three neuron types. Individual IPSCs (grey traces) and average IPSCs (color-coded traces) are shown superimposed. Bath application of 10 µM SR59931 blocked IPSCs in three slow-firing ChAT cells. Right, summary plots show peak amplitudes of evoked IPSCs in PVIs (10 out of 32 cells), ChATs (7 out of 49 cells) and putative glutamatergic cells (5 out of 23 cells). Open circles are individual data points, filled circles are mean values with lines representing ± SEM; **p≤0.01, Mann-Whitney U test for pair-wise comparisons between PVIs vs putative glutamatergic cells and ChATs vs glutamatergic cells. ***p<0.001 for pair-wise comparisons between PVIs vs putative glutamatergic cells and PVIs vs ChATs. To compare three data sets for significant differences a Kruskal-Wallis one-way analysis of variance on Ranks was performed.